Probing structures of large protein complexes using zero-length cross-linking
- PMID: 25937394
- PMCID: PMC4628899
- DOI: 10.1016/j.ymeth.2015.04.031
Probing structures of large protein complexes using zero-length cross-linking
Abstract
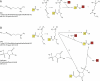
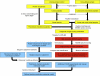
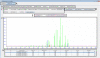
Structural mass spectrometry (MS) is a field with growing applicability for addressing complex biophysical questions regarding proteins and protein complexes. One of the major structural MS approaches involves the use of chemical cross-linking coupled with MS analysis (CX-MS) to identify proximal sites within macromolecules. Identified cross-linked sites can be used to probe novel protein-protein interactions or the derived distance constraints can be used to verify and refine molecular models. This review focuses on recent advances of "zero-length" cross-linking. Zero-length cross-linking reagents do not add any atoms to the cross-linked species due to the lack of a spacer arm. This provides a major advantage in the form of providing more precise distance constraints as the cross-linkable groups must be within salt bridge distances in order to react. However, identification of cross-linked peptides using these reagents presents unique challenges. We discuss recent efforts by our group to minimize these challenges by using multiple cycles of LC-MS/MS analysis and software specifically developed and optimized for identification of zero-length cross-linked peptides. Representative data utilizing our current protocol are presented and discussed.
Keywords: Chemical cross-linking; Cross-link identification; Mass spectrometry; Molecular modeling; Structure; Zero-length cross-linking.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
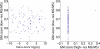





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

