Comparing Platforms for Messenger RNA Expression Profiling of Archival Formalin-Fixed, Paraffin-Embedded Tissues
- PMID: 25937617
- PMCID: PMC4483460
- DOI: 10.1016/j.jmoldx.2015.02.002
Comparing Platforms for Messenger RNA Expression Profiling of Archival Formalin-Fixed, Paraffin-Embedded Tissues
Abstract
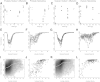
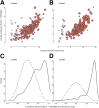
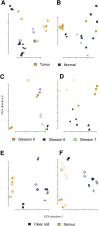
Archival formalin-fixed, paraffin-embedded (FFPE) tissue specimens represent a readily available but largely untapped resource for gene expression profiling-based biomarker discovery. Several technologies have been proposed to cope with the bias from RNA cross-linking and degradation associated with archival specimens to generate data comparable with RNA from fresh-frozen materials. Direct comparison studies of these RNA expression platforms remain rare. We compared two commercially available platforms for RNA expression profiling of archival FFPE specimens from clinical studies of prostate and ovarian cancer: the Affymetrix Human Gene 1.0ST Array following whole-transcriptome amplification using the NuGen WT-Ovation FFPE System V2, and the NanoString nCounter without amplification. For each assay, we profiled 7 prostate and 11 ovarian cancer specimens, with a block age of 4 to 21 years. Both platforms produced gene expression profiles with high sensitivity and reproducibility through technical repeats from FFPE materials. Sensitivity and reproducibility remained high across block age within each cohort. A strong concordance was shown for the transcript expression values for genes detected by both platforms. We showed the biological validity of specific gene signatures generated by both platforms for both cohorts. Our study supports the feasibility of gene expression profiling and large-scale signature validation on archival prostate and ovarian tumor specimens using commercial platforms. These approaches have the potential to aid precision medicine with biomarker discovery and validation.
Copyright © 2015 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Mirnezami R., Nicholson J., Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366:489–491. - PubMed
-
- Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., Louis D.N., Christiani D.C., Settleman J., Haber D.A. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Slamon D.J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. - PubMed
-
- Paik S., Shak S., Tang G., Kim C., Baker J., Cronin M., Baehner F.L., Walker M.G., Watson D., Park T., Hiller W., Fisher E.R., Wickerham D.L., Bryant J., Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. - PubMed
-
- Hoshida Y., Villanueva A., Kobayashi M., Peix J., Chiang D.Y., Camargo A., Gupta S., Moore J., Wrobel M.J., Lerner J., Reich M., Chan J.A., Glickman J.N., Ikeda K., Hashimoto M., Watanabe G., Daidone M.G., Roayaie S., Schwartz M., Thung S., Salvesen H.B., Gabriel S., Mazzaferro V., Bruix J., Friedman S.L., Kumada H., Llovet J.M., Golub T.R. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA090381/CA/NCI NIH HHS/United States
- CA136578/CA/NCI NIH HHS/United States
- R01CA131945/CA/NCI NIH HHS/United States
- 5R01CA142832-05/CA/NCI NIH HHS/United States
- 5R01CA141298/CA/NCI NIH HHS/United States
- 1RC4CA156551-01/CA/NCI NIH HHS/United States
- R01 CA131945/CA/NCI NIH HHS/United States
- P50CA90381/CA/NCI NIH HHS/United States
- R01 CA142832/CA/NCI NIH HHS/United States
- CA090381/CA/NCI NIH HHS/United States
- R01 CA136578/CA/NCI NIH HHS/United States
- RC4 CA156551/CA/NCI NIH HHS/United States
- R01 CA141298/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

