Reverse Engineering of Genome-wide Gene Regulatory Networks from Gene Expression Data
- PMID: 25937810
- PMCID: PMC4412962
- DOI: 10.2174/1389202915666141110210634
Reverse Engineering of Genome-wide Gene Regulatory Networks from Gene Expression Data
Abstract
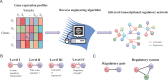
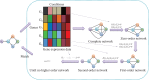
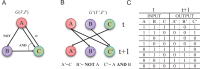
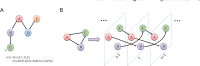
Transcriptional regulation plays vital roles in many fundamental biological processes. Reverse engineering of genome-wide regulatory networks from high-throughput transcriptomic data provides a promising way to characterize the global scenario of regulatory relationships between regulators and their targets. In this review, we summarize and categorize the main frameworks and methods currently available for inferring transcriptional regulatory networks from microarray gene expression profiling data. We overview each of strategies and introduce representative methods respectively. Their assumptions, advantages, shortcomings, and possible improvements and extensions are also clarified and commented.
Keywords: Computational model; Gene expression data; Genome-wide inference; Reverse engineering; Transcriptional regulatory network.
Figures

Similar articles
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
Computational and experimental approaches for modeling gene regulatory networks.Curr Pharm Des. 2007;13(14):1415-36. doi: 10.2174/138161207780765945. Curr Pharm Des. 2007. PMID: 17504165 Review.
-
An Overview of NCA-Based Algorithms for Transcriptional Regulatory Network Inference.Microarrays (Basel). 2015 Nov 16;4(4):596-617. doi: 10.3390/microarrays4040596. Microarrays (Basel). 2015. PMID: 27600242 Free PMC article. Review.
-
Transcription regulatory networks in Caenorhabditis elegans inferred through reverse-engineering of gene expression profiles constitute biological hypotheses for metazoan development.Mol Biosyst. 2009 Dec;5(12):1817-30. doi: 10.1039/B908108a. Epub 2009 Jul 17. Mol Biosyst. 2009. PMID: 19763340
-
Inferring active regulatory networks from gene expression data using a combination of prior knowledge and enrichment analysis.BMC Bioinformatics. 2016 Jun 6;17 Suppl 5(Suppl 5):181. doi: 10.1186/s12859-016-1040-7. BMC Bioinformatics. 2016. PMID: 27295045 Free PMC article.
Cited by
-
Diurnal Transcriptome and Gene Network Represented through Sparse Modeling in Brachypodium distachyon.Front Plant Sci. 2017 Nov 28;8:2055. doi: 10.3389/fpls.2017.02055. eCollection 2017. Front Plant Sci. 2017. PMID: 29234348 Free PMC article.
-
Candidate genes in gastric cancer identified by constructing a weighted gene co-expression network.PeerJ. 2018 May 4;6:e4692. doi: 10.7717/peerj.4692. eCollection 2018. PeerJ. 2018. PMID: 29740513 Free PMC article.
-
A Bayesian data fusion based approach for learning genome-wide transcriptional regulatory networks.BMC Bioinformatics. 2020 May 29;21(1):219. doi: 10.1186/s12859-020-3510-1. BMC Bioinformatics. 2020. PMID: 32471360 Free PMC article.
-
Nonlinear expression patterns and multiple shifts in gene network interactions underlie robust phenotypic change in Drosophila melanogaster selected for night sleep duration.PLoS Comput Biol. 2023 Aug 10;19(8):e1011389. doi: 10.1371/journal.pcbi.1011389. eCollection 2023 Aug. PLoS Comput Biol. 2023. PMID: 37561813 Free PMC article.
-
Global Transcriptome Analysis of Combined Abiotic Stress Signaling Genes Unravels Key Players in Oryza sativa L.: An In silico Approach.Front Plant Sci. 2017 May 15;8:759. doi: 10.3389/fpls.2017.00759. eCollection 2017. Front Plant Sci. 2017. PMID: 28555143 Free PMC article.
References
-
- Spitz F., Furlong E.E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13(9):613–626. - PubMed
-
- Chen K., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8(2):93–103. - PubMed
-
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108(4):439–451. - PubMed
-
- Babu M.M., Luscombe N.M., Aravind L., Gerstein M., Teichmann S.A. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14(3):283–291. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
