Extracting research-quality phenotypes from electronic health records to support precision medicine
- PMID: 25937834
- PMCID: PMC4416392
- DOI: 10.1186/s13073-015-0166-y
Extracting research-quality phenotypes from electronic health records to support precision medicine
Abstract
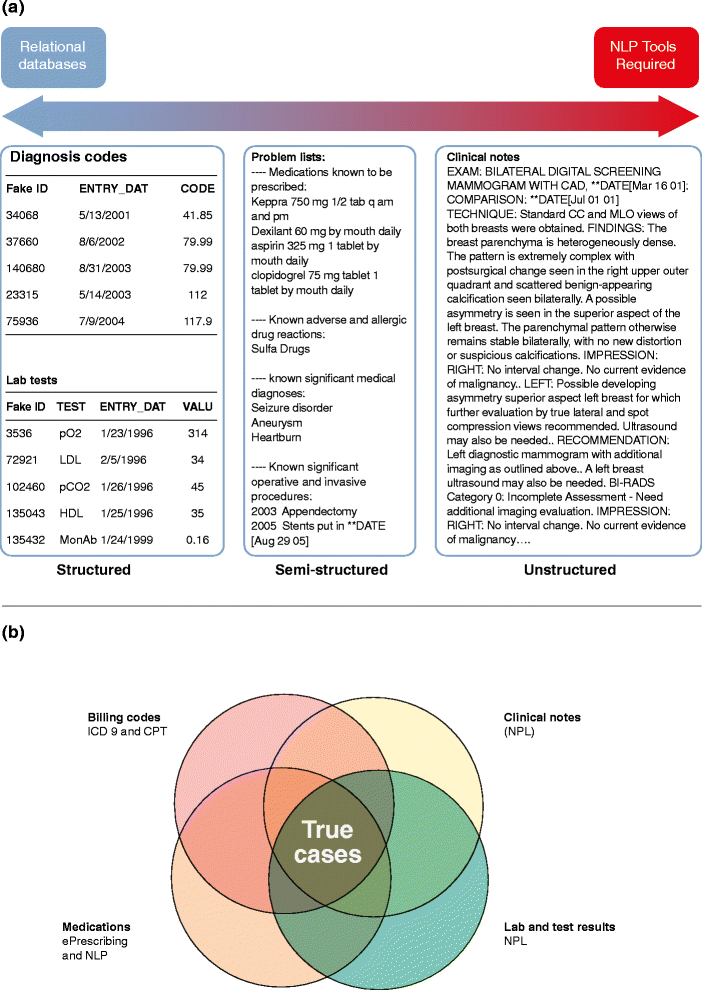
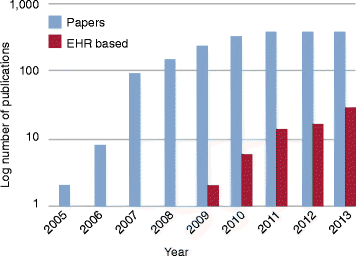
The convergence of two rapidly developing technologies - high-throughput genotyping and electronic health records (EHRs) - gives scientists an unprecedented opportunity to utilize routine healthcare data to accelerate genomic discovery. Institutions and healthcare systems have been building EHR-linked DNA biobanks to enable such a vision. However, the precise extraction of detailed disease and drug-response phenotype information hidden in EHRs is not an easy task. EHR-based studies have successfully replicated known associations, made new discoveries for diseases and drug response traits, rapidly contributed cases and controls to large meta-analyses, and demonstrated the potential of EHRs for broad-based phenome-wide association studies. In this review, we summarize the advantages and challenges of repurposing EHR data for genetic research. We also highlight recent notable studies and novel approaches to provide an overview of advanced EHR-based phenotyping.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

