TermGenie - a web-application for pattern-based ontology class generation
- PMID: 25937883
- PMCID: PMC4417543
- DOI: 10.1186/2041-1480-5-48
TermGenie - a web-application for pattern-based ontology class generation
Abstract
Background: Biological ontologies are continually growing and improving from requests for new classes (terms) by biocurators. These ontology requests can frequently create bottlenecks in the biocuration process, as ontology developers struggle to keep up, while manually processing these requests and create classes.
Results: TermGenie allows biocurators to generate new classes based on formally specified design patterns or templates. The system is web-based and can be accessed by any authorized curator through a web browser. Automated rules and reasoning engines are used to ensure validity, uniqueness and relationship to pre-existing classes. In the last 4 years the Gene Ontology TermGenie generated 4715 new classes, about 51.4% of all new classes created. The immediate generation of permanent identifiers proved not to be an issue with only 70 (1.4%) obsoleted classes.
Conclusion: TermGenie is a web-based class-generation system that complements traditional ontology development tools. All classes added through pre-defined templates are guaranteed to have OWL equivalence axioms that are used for automatic classification and in some cases inter-ontology linkage. At the same time, the system is simple and intuitive and can be used by most biocurators without extensive training.
Keywords: Class generation; Ontology.
Figures
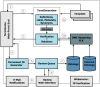

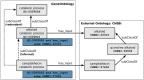

References
-
- Gene ontology developer documentationhttp://geneontology.org/page/development
-
- jQuery javascript libraryhttp://jquery.com
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

