Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance
- PMID: 25938965
- PMCID: PMC4463611
- DOI: 10.3390/ijms16059654
Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance
Abstract
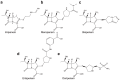
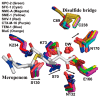
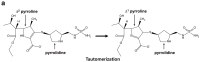
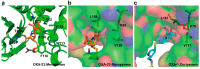
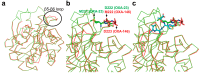
Carbapenems (imipenem, meropenem, biapenem, ertapenem, and doripenem) are β-lactam antimicrobial agents. Because carbapenems have the broadest spectra among all β-lactams and are primarily used to treat infections by multi-resistant Gram-negative bacteria, the emergence and spread of carbapenemases became a major public health concern. Carbapenemases are the most versatile family of β-lactamases that are able to hydrolyze carbapenems and many other β-lactams. According to the dependency of divalent cations for enzyme activation, carbapenemases can be divided into metallo-carbapenemases (zinc-dependent class B) and non-metallo-carbapenemases (zinc-independent classes A, C, and D). Many studies have provided various carbapenemase structures. Here we present a comprehensive and systematic review of three-dimensional structures of carbapenemase-carbapenem complexes as well as those of carbapenemases. We update recent studies in understanding the enzymatic mechanism of each class of carbapenemase, and summarize structural insights about regions and residues that are important in acquiring the carbapenemase activity.
Figures

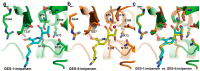
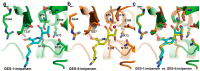
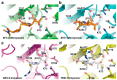




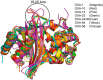


References
-
- Lee J.H., Lee S.H. Carbapenem resistance in gram-negative pathogens: Emerging non-metallo-carbapenemases. Res. J. Microbiol. 2006;1:1–22. doi: 10.3923/jm.2006.1.22. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

