GXD: a community resource of mouse Gene Expression Data
- PMID: 25939429
- PMCID: PMC4534488
- DOI: 10.1007/s00335-015-9563-1
GXD: a community resource of mouse Gene Expression Data
Abstract
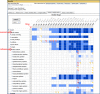
The Gene Expression Database (GXD) is an extensive, easily searchable, and freely available database of mouse gene expression information (www.informatics.jax.org/expression.shtml). GXD was developed to foster progress toward understanding the molecular basis of human development and disease. GXD contains information about when and where genes are expressed in different tissues in the mouse, especially during the embryonic period. GXD collects different types of expression data from wild-type and mutant mice, including RNA in situ hybridization, immunohistochemistry, RT-PCR, and northern and western blot results. The GXD curators read the scientific literature and enter the expression data from those papers into the database. GXD also acquires expression data directly from researchers, including groups doing large-scale expression studies. GXD currently contains nearly 1.5 million expression results for over 13,900 genes. In addition, it has over 265,000 images of expression data, allowing users to retrieve the primary data and interpret it themselves. By being an integral part of the larger Mouse Genome Informatics (MGI) resource, GXD's expression data are combined with other genetic, functional, phenotypic, and disease-oriented data. This allows GXD to provide tools for researchers to evaluate expression data in the larger context, search by a wide variety of biologically and biomedically relevant parameters, and discover new data connections to help in the design of new experiments. Thus, GXD can provide researchers with critical insights into the functions of genes and the molecular mechanisms of development, differentiation, and disease.
Figures



References
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
-
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nürnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dollé P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

