An automated imaging system for radiation biodosimetry
- PMID: 25939519
- PMCID: PMC4479970
- DOI: 10.1002/jemt.22512
An automated imaging system for radiation biodosimetry
Abstract
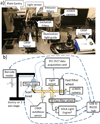
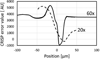
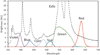
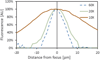
We describe here an automated imaging system developed at the Center for High Throughput Minimally Invasive Radiation Biodosimetry. The imaging system is built around a fast, sensitive sCMOS camera and rapid switchable LED light source. It features complete automation of all the steps of the imaging process and contains built-in feedback loops to ensure proper operation. The imaging system is intended as a back end to the RABiT-a robotic platform for radiation biodosimetry. It is intended to automate image acquisition and analysis for four biodosimetry assays for which we have developed automated protocols: The Cytokinesis Blocked Micronucleus assay, the γ-H2AX assay, the Dicentric assay (using PNA or FISH probes) and the RABiT-BAND assay.
Keywords: dicentrics; fluorescence microscopy; mBAND; micronuclei; sCMOS; γ-H2AX.
© 2015 Wiley Periodicals, Inc.
Conflict of interest statement
Some of the reagents, equipment and plasticware used in this work were purchased through Fisher Scientific. At the time of writing this manuscript GG owns 90 shares of Thermo Fisher Scientific stock. The Authors report no other potential conflicts.
Figures






References
-
- Böcker W, Iliakis G. Computational Methods for Analysis of Foci: Validation for Radiation-Induced γ-H2AX Foci in Human Cells. Radiation Research. 2006;165(1):113–124. - PubMed
-
- Bueno G, Déniz O, Fernández-Carrobles MDM, Vállez N, Salido J. An automated system for whole microscopic image acquisition and analysis. Microscopy Research and Technique. 2014;77(9):697–713. - PubMed
-
- Chen Y, Zhang J, Wang H, Garty G, Xu Y, Lyulko OV, Turner HC, Randers-Pehrson G, Simaan N, Yao YL, Brenner DJ. Development of a Robotically-based Automated Biodosimetry Tool for High-throughput Radiological Triage. International Journal of Biomechatronics and Biomedical Robotics. 2010;1(2):115–125.
-
- Chudoba I, Hickmann G, Friedrich T, Jauch A, Kozlowski P, Senger G. mBAND: a high resolution multicolor banding technique for the detection of complex intrachromosomal aberrations. Cytogenet Genome Res. 2004;104(1–4):390–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

