The Function of Embryonic Stem Cell-expressed RAS (E-RAS), a Unique RAS Family Member, Correlates with Its Additional Motifs and Its Structural Properties
- PMID: 25940089
- PMCID: PMC4505495
- DOI: 10.1074/jbc.M115.640607
The Function of Embryonic Stem Cell-expressed RAS (E-RAS), a Unique RAS Family Member, Correlates with Its Additional Motifs and Its Structural Properties
Abstract
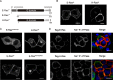
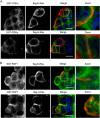
E-RAS is a member of the RAS family specifically expressed in embryonic stem cells, gastric tumors, and hepatic stellate cells. Unlike classical RAS isoforms (H-, N-, and K-RAS4B), E-RAS has, in addition to striking and remarkable sequence deviations, an extended 38-amino acid-long unique N-terminal region with still unknown functions. We investigated the molecular mechanism of E-RAS regulation and function with respect to its sequence and structural features. We found that N-terminal extension of E-RAS is important for E-RAS signaling activity. E-RAS protein most remarkably revealed a different mode of effector interaction as compared with H-RAS, which correlates with deviations in the effector-binding site of E-RAS. Of all these residues, tryptophan 79 (arginine 41 in H-RAS), in the interswitch region, modulates the effector selectivity of RAS proteins from H-RAS to E-RAS features.
Keywords: E-RAS; H-RAS; RAS protein; Raf kinase; effector selection; embryonic stem cell-expressed RAS; phosphatidylinositide 3-kinase (PI 3-kinase); phosphatidylinositol kinase (PI kinase); small GTPase; specificity determining residues.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

