From the core to beyond the margin: a genomic picture of glioblastoma intratumor heterogeneity
- PMID: 25940437
- PMCID: PMC4494925
- DOI: 10.18632/oncotarget.3297
From the core to beyond the margin: a genomic picture of glioblastoma intratumor heterogeneity
Erratum in
-
Correction: From the core to beyond the margin: a genomic picture of glioblastoma intratumor heterogeneity.Oncotarget. 2016 Oct 11;7(41):67685. doi: 10.18632/oncotarget.12550. Oncotarget. 2016. PMID: 28049185 Free PMC article. No abstract available.
Abstract
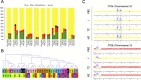
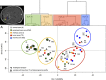
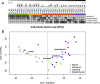
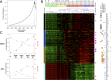
Glioblastoma (GB) is a highly invasive primary brain tumor that almost systematically recurs despite aggressive therapies. One of the most challenging problems in therapy of GB is its extremely complex and heterogeneous molecular biology. To explore this heterogeneity, we performed a genome-wide integrative screening of three molecular levels: genome, transcriptome, and methylome. We analyzed tumor biopsies obtained by neuro-navigation in four distinct areas for 10 GB patients (necrotic zone, tumor zone, interface, and peripheral brain zone). We classified samples and deciphered a key genes signature of intratumor heterogeneity by Principal Component Analysis and Weighted Gene Co-expression Network Analysis. At the genome level, we identified common GB copy number alterations and but a strong interindividual molecular heterogeneity. Transcriptome analysis highlighted a pronounced intratumor architecture reflecting the surgical sampling plan of the study and identified gene modules associated with hallmarks of cancer. We provide a signature of key cancer-heterogeneity genes highly associated with the intratumor spatial gradient and show that it is enriched in genes with correlation between methylation and expression levels. Our study confirms that GBs are molecularly highly diverse and that a single tumor can harbor different transcriptional GB subtypes depending on its spatial architecture.
Keywords: glioblastoma; integrative functional genomics; intratumor heterogeneity; invasion.
Conflict of interest statement
The author(s) declare that they have no competing interests.
Figures

References
-
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. - PubMed
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

