The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids
- PMID: 25940626
- PMCID: PMC4477650
- DOI: 10.1093/nar/gkv406
The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids
Abstract
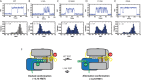
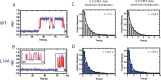
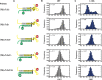
Telomerase is an enzyme that adds repetitive DNA sequences to the ends of chromosomes and consists of two main subunits: the telomerase reverse transcriptase (TERT) protein and an associated telomerase RNA (TER). The telomerase essential N-terminal (TEN) domain is a conserved region of TERT proposed to mediate DNA substrate interactions. Here, we have employed single molecule telomerase binding assays to investigate the function of the TEN domain. Our results reveal telomeric DNA substrates bound to telomerase exhibit a dynamic equilibrium between two states: a docked conformation and an alternative conformation. The relative stabilities of the docked and alternative states correlate with the number of basepairs that can be formed between the DNA substrate and the RNA template, with more basepairing favoring the docked state. The docked state is further buttressed by the TEN domain and mutations within the TEN domain substantially alter the DNA substrate structural equilibrium. We propose a model in which the TEN domain stabilizes short RNA-DNA duplexes in the active site of the enzyme, promoting the docked state to augment telomerase processivity.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Palm W., de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. - PubMed
-
- Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. - PubMed
-
- Vulliamy T.J., Dokal I. Dyskeratosis congenita: the diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2008;90:122–130. - PubMed
-
- Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

