Stabilization of G protein-coupled receptors by point mutations
- PMID: 25941489
- PMCID: PMC4403299
- DOI: 10.3389/fphar.2015.00082
Stabilization of G protein-coupled receptors by point mutations
Abstract
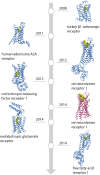
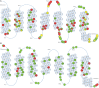
G protein-coupled receptors (GPCRs) are flexible integral membrane proteins involved in transmembrane signaling. Their involvement in many physiological processes makes them interesting targets for drug development. Determination of the structure of these receptors will help to design more specific drugs, however, their structural characterization has so far been hampered by the low expression and their inherent instability in detergents which made protein engineering indispensable for structural and biophysical characterization. Several approaches to stabilize the receptors in a particular conformation have led to breakthroughs in GPCR structure determination. These include truncations of the flexible regions, stabilization by antibodies and nanobodies, fusion partners, high affinity and covalently bound ligands as well as conformational stabilization by mutagenesis. In this review we focus on stabilization of GPCRs by insertion of point mutations, which lead to increased conformational and thermal stability as well as improved expression levels. We summarize existing mutagenesis strategies with different coverage of GPCR sequence space and depth of information, design and transferability of mutations and the molecular basis for stabilization. We also discuss whether mutations alter the structure and pharmacological properties of GPCRs.
Keywords: G protein-coupled receptors; GPCRs; alanine scanning; conformational thermostabilization; pharmacology; protein engineering; stabilizing mutations.
Figures



References
-
- Baker J. G., Proudman R. G. W., Tate C. G. (2011). The pharmacological effects of the thermostabilising (m23) mutations and intra and extracellular (β36) deletions essential for crystallisation of the turkey β-adrenoceptor. Naunyn. Schmiedebergs. Arch. Pharmacol. 384, 71–91. 10.1007/s00210-011-0648-4 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

