Ultra-high density intra-specific genetic linkage maps accelerate identification of functionally relevant molecular tags governing important agronomic traits in chickpea
- PMID: 25942004
- PMCID: PMC5386344
- DOI: 10.1038/srep09468
Ultra-high density intra-specific genetic linkage maps accelerate identification of functionally relevant molecular tags governing important agronomic traits in chickpea
Abstract
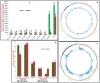
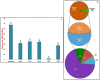
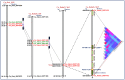
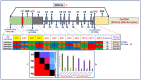
We discovered 26785 and 16573 high-quality SNPs differentiating two parental genotypes of a RIL mapping population using reference desi and kabuli genome-based GBS assay. Of these, 3625 and 2177 SNPs have been integrated into eight desi and kabuli chromosomes, respectively in order to construct ultra-high density (0.20-0.37 cM) intra-specific chickpea genetic linkage maps. One of these constructed high-resolution genetic map has potential to identify 33 major genomic regions harbouring 35 robust QTLs (PVE: 17.9-39.7%) associated with three agronomic traits, which were mapped within <1 cM mean marker intervals on desi chromosomes. The extended LD (linkage disequilibrium) decay (~15 cM) in chromosomes of genetic maps have encouraged us to use a rapid integrated approach (comparative QTL mapping, QTL-region specific haplotype/LD-based trait association analysis, expression profiling and gene haplotype-based association mapping) rather than a traditional QTL map-based cloning method to narrow-down one major seed weight (SW) robust QTL region. It delineated favourable natural allelic variants and superior haplotype-containing one seed-specific candidate embryo defective gene regulating SW in chickpea. The ultra-high-resolution genetic maps, QTLs/genes and alleles/haplotypes-related genomic information generated and integrated strategy for rapid QTL/gene identification developed have potential to expedite genomics-assisted breeding applications in crop plants, including chickpea for their genetic enhancement.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Jukanti A. K. et al. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br. J. Nutr. 108, S11–26 (2012). - PubMed
-
- Arumuganathan K. & Earle E. D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–219 (1991).
-
- Kumar A., Choudhary A. K., Solanki R. K. & Pratap A. Towards marker-assisted selection in pulses: a review. Plant Breed. 130, 297–313 (2011).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

