A Novel Type II NAD+-Specific Isocitrate Dehydrogenase from the Marine Bacterium Congregibacter litoralis KT71
- PMID: 25942017
- PMCID: PMC4420465
- DOI: 10.1371/journal.pone.0125229
A Novel Type II NAD+-Specific Isocitrate Dehydrogenase from the Marine Bacterium Congregibacter litoralis KT71
Abstract
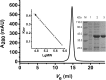
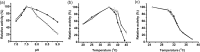
In most living organisms, isocitrate dehydrogenases (IDHs) convert isocitrate into ɑ-ketoglutarate (ɑ-KG). Phylogenetic analyses divide the IDH protein family into two subgroups: types I and II. Based on cofactor usage, IDHs are either NAD+-specific (NAD-IDH) or NADP+-specific (NADP-IDH); NADP-IDH evolved from NAD-IDH. Type I IDHs include NAD-IDHs and NADP-IDHs; however, no type II NAD-IDHs have been reported to date. This study reports a novel type II NAD-IDH from the marine bacterium Congregibacter litoralis KT71 (ClIDH, GenBank accession no. EAQ96042). His-tagged recombinant ClIDH was produced in Escherichia coli and purified; the recombinant enzyme was NAD+-specific and showed no detectable activity with NADP+. The Km values of the enzyme for NAD+ were 262.6±7.4 μM or 309.1±11.2 μM with Mg2+ or Mn2+ as the divalent cation, respectively. The coenzyme specificity of a ClIDH Asp487Arg/Leu488His mutant was altered, and the preference of the mutant for NADP+ was approximately 24-fold higher than that for NAD+, suggesting that ClIDH is an NAD+-specific ancestral enzyme in the type II IDH subgroup. Gel filtration and analytical ultracentrifugation analyses revealed the homohexameric structure of ClIDH, which is the first IDH hexamer discovered thus far. A 163-amino acid segment of CIIDH is essential to maintain its polymerization structure and activity, as a truncated version lacking this region forms a non-functional monomer. ClIDH was dependent on divalent cations, the most effective being Mn2+. The maximal activity of purified recombinant ClIDH was achieved at 35°C and pH 7.5, and a heat inactivation experiment showed that a 20-min incubation at 33°C caused a 50% loss of ClIDH activity. The discovery of a NAD+-specific, type II IDH fills a gap in the current classification of IDHs, and sheds light on the evolution of type II IDHs.
Conflict of interest statement
Figures




References
-
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010; 17(3): 225–34. 10.1016/j.ccr.2010.01.020 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

