The RCSB PDB "Molecule of the Month": Inspiring a Molecular View of Biology
- PMID: 25942442
- PMCID: PMC4420264
- DOI: 10.1371/journal.pbio.1002140
The RCSB PDB "Molecule of the Month": Inspiring a Molecular View of Biology
Abstract
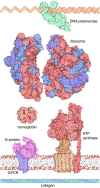
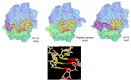
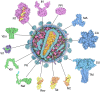
The Research Collaboratory for Structural Bioinformatics (RCSB) Molecule of the Month series provides a curated introduction to the 3-D biomolecular structures available in the Protein Data Bank archive and the tools that are available at the RCSB website for accessing and exploring them. A variety of educational materials, such as articles, videos, posters, hands-on activities, lesson plans, and curricula, build on this series for use in a variety of educational settings as a general introduction to key topics, such as enzyme action, protein synthesis, and viruses. The series and associated educational materials are freely available at www.rcsb.org.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Martz E. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem Sci 2002; 27: 107–109. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

