Optimization of genomic selection training populations with a genetic algorithm
- PMID: 25943105
- PMCID: PMC4422310
- DOI: 10.1186/s12711-015-0116-6
Optimization of genomic selection training populations with a genetic algorithm
Abstract
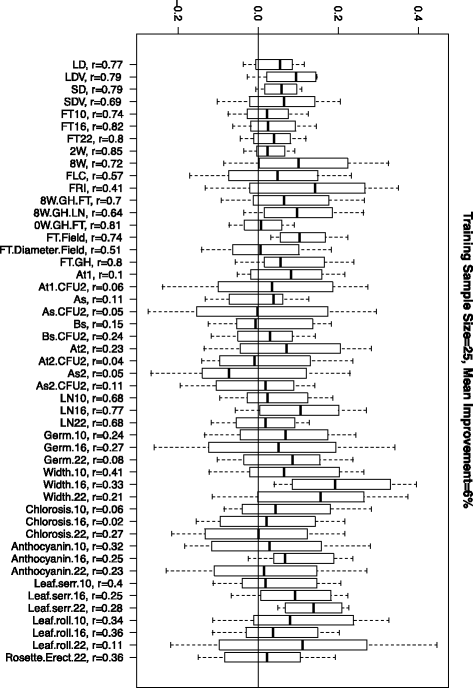
In this article, we imagine a breeding scenario with a population of individuals that have been genotyped but not phenotyped. We derived a computationally efficient statistic that uses this genetic information to measure the reliability of genomic estimated breeding values (GEBV) for a given set of individuals (test set) based on a training set of individuals. We used this reliability measure with a genetic algorithm scheme to find an optimized training set from a larger set of candidate individuals. This subset was phenotyped to create the training set that was used in a genomic selection model to estimate GEBV in the test set. Our results show that, compared to a random sample of the same size, the use of a set of individuals selected by our method improved accuracies. We implemented the proposed training selection methodology on four sets of data on Arabidopsis, wheat, rice and maize. This dynamic model building process that takes genotypes of the individuals in the test sample into account while selecting the training individuals improves the performance of genomic selection models.
Figures




References
-
- Heslot N, Yang HP, Sorrells ME, Jannink JL. Genomic selection in plant breeding: a comparison of models. Crop Sci. 2012;52:146–60. doi: 10.2135/cropsci2011.06.0297. - DOI
-
- Rincent R, Laloë D. Nicolas S, Altmann T, Brunel D, Revilla P, et al. Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: Comparison of methods in two diverse groups of maize inbreds (zea mays l.) Genetics. 2012;192:715–28. doi: 10.1534/genetics.112.141473. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

