Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N
- PMID: 25943375
- PMCID: PMC4441053
- DOI: 10.1101/gad.259432.115
Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N
Abstract
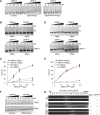
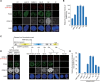
Specific recognition of centromere-specific histone variant CENP-A-containing chromatin by CENP-N is an essential process in the assembly of the kinetochore complex at centromeres prior to mammalian cell division. However, the mechanisms of CENP-N recruitment to centromeres/kinetochores remain unknown. Here, we show that a CENP-A-specific RG loop (Arg80/Gly81) plays an essential and dual regulatory role in this process. The RG loop assists the formation of a compact "ladder-like" structure of CENP-A chromatin, concealing the loop and thus impairing its role in recruiting CENP-N. Upon G1/S-phase transition, however, centromeric chromatin switches from the compact to an open state, enabling the now exposed RG loop to recruit CENP-N prior to cell division. Our results provide the first insights into the mechanisms by which the recruitment of CENP-N is regulated by the structural transitions between compaction and relaxation of centromeric chromatin during the cell cycle.
Keywords: CENP-A; CENP-N; RG loop; cell cycle; chromosome congression; higher-order chromatin structure.
© 2015 Fang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
