HMMER web server: 2015 update
- PMID: 25943547
- PMCID: PMC4489315
- DOI: 10.1093/nar/gkv397
HMMER web server: 2015 update
Abstract
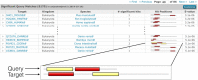
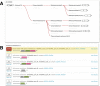
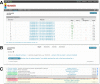
The HMMER website, available at http://www.ebi.ac.uk/Tools/hmmer/, provides access to the protein homology search algorithms found in the HMMER software suite. Since the first release of the website in 2011, the search repertoire has been expanded to include the iterative search algorithm, jackhmmer. The continued growth of the target sequence databases means that traditional tabular representations of significant sequence hits can be overwhelming to the user. Consequently, additional ways of presenting homology search results have been developed, allowing them to be summarised according to taxonomic distribution or domain architecture. The taxonomy and domain architecture representations can be used in combination to filter the results according to the needs of a user. Searches can also be restricted prior to submission using a new taxonomic filter, which not only ensures that the results are specific to the requested taxonomic group, but also improves search performance. The repertoire of profile hidden Markov model libraries, which are used for annotation of query sequences with protein families and domains, has been expanded to include the libraries from CATH-Gene3D, PIRSF, Superfamily and TIGRFAMs. Finally, we discuss the relocation of the HMMER webserver to the European Bioinformatics Institute and the potential impact that this will have.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

