Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy
- PMID: 25943887
- PMCID: PMC4468790
- DOI: 10.1007/s00401-015-1429-9
Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy
Abstract
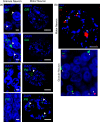
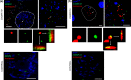
GGGGCC repeat expansions of C9ORF72 represent the most common genetic variant of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia. We and others have proposed that RNA transcribed from the repeat sequence is toxic via sequestration of RNA-binding factors. Both GGGGCC-repeat (sense) and CCCCGG-repeat (antisense) molecules are detectable by fluorescence in situ hybridisation as RNA foci, but their relative expression pattern within the CNS and contribution to disease has not been determined. Blinded examination of CNS biosamples from ALS patients with a repeat expansion of C9ORF72 showed that antisense foci are present at a significantly higher frequency in cerebellar Purkinje neurons and motor neurons, whereas sense foci are present at a significantly higher frequency in cerebellar granule neurons. Consistent with this, inclusions containing sense or antisense derived dipeptide repeat proteins were present at significantly higher frequency in cerebellar granule neurons or motor neurons, respectively. Immunohistochemistry and UV-crosslinking studies showed that sense and antisense RNA molecules share similar interactions with SRSF2, hnRNP K, hnRNP A1, ALYREF, and hnRNP H/F. Together these data suggest that, although sense and antisense RNA molecules might be expected to be equally toxic via their shared protein binding partners, distinct patterns of expression in various CNS neuronal populations could lead to relative differences in their contribution to the pathogenesis of neuronal injury. Moreover in motor neurons, which are the primary target of pathology in ALS, the presence of antisense foci (χ (2), p < 0.00001) but not sense foci (χ (2), p = 0.75) correlated with mislocalisation of TDP-43, which is the hallmark of ALS neurodegeneration. This has implications for translational approaches to C9ORF72 disease, and furthermore interacting RNA-processing factors and transcriptional activators responsible for antisense versus sense transcription might represent novel therapeutic targets.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- MR/L016451/1/MRC_/Medical Research Council/United Kingdom
- 089701/WT_/Wellcome Trust/United Kingdom
- MR/K000039/1/MRC_/Medical Research Council/United Kingdom
- G1100540/MRC_/Medical Research Council/United Kingdom
- G0800380/MRC_/Medical Research Council/United Kingdom
- G0900652/MRC_/Medical Research Council/United Kingdom
- MR/K003771/1/MRC_/Medical Research Council/United Kingdom
- G0400074/MRC_/Medical Research Council/United Kingdom
- COOPER-KNOCK/FEB12/942-795/MNDA_/Motor Neurone Disease Association/United Kingdom
- G0 800380/MRC_/Medical Research Council/United Kingdom
- G0502157/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

