Autophagic recycling plays a central role in maize nitrogen remobilization
- PMID: 25944100
- PMCID: PMC4456646
- DOI: 10.1105/tpc.15.00158
Autophagic recycling plays a central role in maize nitrogen remobilization
Abstract
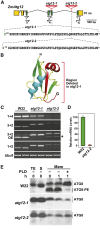
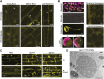
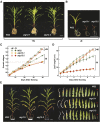
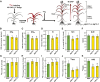
Autophagy is a primary route for nutrient recycling in plants by which superfluous or damaged cytoplasmic material and organelles are encapsulated and delivered to the vacuole for breakdown. Central to autophagy is a conjugation pathway that attaches AUTOPHAGY-RELATED8 (ATG8) to phosphatidylethanolamine, which then coats emerging autophagic membranes and helps with cargo recruitment, vesicle enclosure, and subsequent vesicle docking with the tonoplast. A key component in ATG8 function is ATG12, which promotes lipidation upon its attachment to ATG5. Here, we fully defined the maize (Zea mays) ATG system transcriptionally and characterized it genetically through atg12 mutants that block ATG8 modification. atg12 plants have compromised autophagic transport as determined by localization of a YFP-ATG8 reporter and its vacuolar cleavage during nitrogen or fixed-carbon starvation. Phenotypic analyses showed that atg12 plants are phenotypically normal and fertile when grown under nutrient-rich conditions. However, when nitrogen-starved, seedling growth is severely arrested, and as the plants mature, they show enhanced leaf senescence and stunted ear development. Nitrogen partitioning studies revealed that remobilization is impaired in atg12 plants, which significantly decreases seed yield and nitrogen-harvest index. Together, our studies demonstrate that autophagy, while nonessential, becomes critical during nitrogen stress and severely impacts maize productivity under suboptimal field conditions.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures




Similar articles
-
The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability.Plant Physiol. 2009 Jan;149(1):220-34. doi: 10.1104/pp.108.126714. Epub 2008 Sep 12. Plant Physiol. 2009. PMID: 18790996 Free PMC article.
-
ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci.Plant J. 2010 May;62(3):483-93. doi: 10.1111/j.1365-313X.2010.04166.x. Epub 2010 Feb 3. Plant J. 2010. PMID: 20136727
-
The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana.Genetics. 2008 Mar;178(3):1339-53. doi: 10.1534/genetics.107.086199. Epub 2008 Feb 3. Genetics. 2008. PMID: 18245858 Free PMC article.
-
Autophagy: a multifaceted intracellular system for bulk and selective recycling.Trends Plant Sci. 2012 Sep;17(9):526-37. doi: 10.1016/j.tplants.2012.05.006. Epub 2012 Jun 11. Trends Plant Sci. 2012. PMID: 22694835 Review.
-
Autophagy, plant senescence, and nutrient recycling.J Exp Bot. 2014 Jul;65(14):3799-811. doi: 10.1093/jxb/eru039. Epub 2014 Mar 31. J Exp Bot. 2014. PMID: 24687977 Review.
Cited by
-
Autophagy Dances with Phytohormones upon Multiple Stresses.Plants (Basel). 2020 Aug 15;9(8):1038. doi: 10.3390/plants9081038. Plants (Basel). 2020. PMID: 32824209 Free PMC article. Review.
-
Genome of Paspalum vaginatum and the role of trehalose mediated autophagy in increasing maize biomass.Nat Commun. 2022 Dec 13;13(1):7731. doi: 10.1038/s41467-022-35507-8. Nat Commun. 2022. PMID: 36513676 Free PMC article.
-
Monitoring Autophagy in Rice With GFP-ATG8 Marker Lines.Front Plant Sci. 2022 Apr 25;13:866367. doi: 10.3389/fpls.2022.866367. eCollection 2022. Front Plant Sci. 2022. PMID: 35548298 Free PMC article.
-
Autophagic pathway contributes to low-nitrogen tolerance by optimizing nitrogen uptake and utilization in tomato.Hortic Res. 2022 Mar 23;9:uhac068. doi: 10.1093/hr/uhac068. eCollection 2022. Hortic Res. 2022. PMID: 35669705 Free PMC article.
-
Genome-wide association analysis for grain moisture content and dehydration rate on maize hybrids.Mol Breed. 2023 Jan 13;43(1):5. doi: 10.1007/s11032-022-01349-x. eCollection 2023 Jan. Mol Breed. 2023. PMID: 37312866 Free PMC article.
References
-
- Abendroth L.J., Elmore R.W., Boyer M.J., Marlay S.K. (2011). Corn Growth and Development. (Ames, IA: Iowa State University Extension; ).
-
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122. - PubMed
-
- Avila-Ospina L., Moison M., Yoshimoto K., Masclaux-Daubresse C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65: 3799–3811. - PubMed
-
- Chardon F., Noël V., Masclaux-Daubresse C. (2012). Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J. Exp. Bot. 63: 3401–3412. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

