Indole Glucosinolate Biosynthesis Limits Phenylpropanoid Accumulation in Arabidopsis thaliana
- PMID: 25944103
- PMCID: PMC4456644
- DOI: 10.1105/tpc.15.00127
Indole Glucosinolate Biosynthesis Limits Phenylpropanoid Accumulation in Arabidopsis thaliana
Abstract
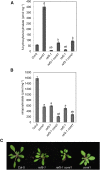
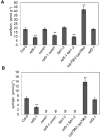
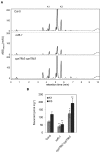
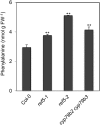
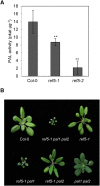
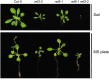
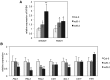
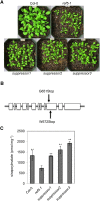
Plants produce an array of metabolites (including lignin monomers and soluble UV-protective metabolites) from phenylalanine through the phenylpropanoid biosynthetic pathway. A subset of plants, including many related to Arabidopsis thaliana, synthesizes glucosinolates, nitrogen- and sulfur-containing secondary metabolites that serve as components of a plant defense system that deters herbivores and pathogens. Here, we report that the Arabidopsis thaliana reduced epidermal fluorescence5 (ref5-1) mutant, identified in a screen for plants with defects in soluble phenylpropanoid accumulation, has a missense mutation in CYP83B1 and displays defects in glucosinolate biosynthesis and in phenylpropanoid accumulation. CYP79B2 and CYP79B3 are responsible for the production of the CYP83B1 substrate indole-3-acetaldoxime (IAOx), and we found that the phenylpropanoid content of cyp79b2 cyp79b3 and ref5-1 cyp79b2 cyp79b3 plants is increased compared with the wild type. These data suggest that levels of IAOx or a subsequent metabolite negatively influence phenylpropanoid accumulation in ref5 and more importantly that this crosstalk is relevant in the wild type. Additional biochemical and genetic evidence indicates that this inhibition impacts the early steps of the phenylpropanoid biosynthetic pathway and restoration of phenylpropanoid accumulation in a ref5-1 med5a/b triple mutant suggests that the function of the Mediator complex is required for the crosstalk.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures
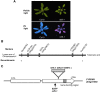
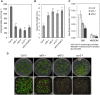
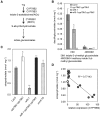
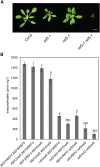
Comment in
-
Metabolic Crosstalk: Interactions between the Phenylpropanoid and Glucosinolate Pathways in Arabidopsis.Plant Cell. 2015 May;27(5):1367. doi: 10.1105/tpc.15.00360. Epub 2015 May 8. Plant Cell. 2015. PMID: 25957387 Free PMC article. No abstract available.
References
-
- Bonawitz N.D., Chapple C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44: 337–363. - PubMed
-
- Bonawitz N.D., Soltau W.L., Blatchley M.R., Powers B.L., Hurlock A.K., Seals L.A., Weng J.-K., Stout J., Chapple C. (2012). REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J. Biol. Chem. 287: 5434–5445. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

