The X-ray structure of Plasmodium falciparum dihydroorotate dehydrogenase bound to a potent and selective N-phenylbenzamide inhibitor reveals novel binding-site interactions
- PMID: 25945708
- PMCID: PMC4427164
- DOI: 10.1107/S2053230X15000989
The X-ray structure of Plasmodium falciparum dihydroorotate dehydrogenase bound to a potent and selective N-phenylbenzamide inhibitor reveals novel binding-site interactions
Abstract
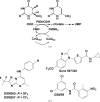
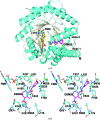
Plasmodium species are protozoan parasites that are the causative agent of malaria. Malaria is a devastating disease, and its treatment and control have been hampered by the propensity of the parasite to become drug-resistant. Dihydroorotate dehydrogenase (DHODH) has been identified as a promising new target for the development of antimalarial agents. Here, the X-ray structure of P. falciparum DHODH bound to a potent and selective N-phenylbenzamide-based inhibitor (DSM59) is described at 2.3 Å resolution. The structure elucidates novel binding-site interactions and shows how conformational flexibility of the enzyme leads to the ability to bind diverse chemical structures with high affinity. This information provides new insight into the design of high-affinity DHODH inhibitors for the treatment of malaria.
Keywords: DSM59; Plasmodium falciparum; dihydroorotate dehydrogenase.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

