Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection
- PMID: 25945999
- PMCID: PMC4422591
- DOI: 10.1371/journal.ppat.1004849
Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection
Erratum in
-
Correction: Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection.PLoS Pathog. 2015 Sep 17;11(9):e1005144. doi: 10.1371/journal.ppat.1005144. eCollection 2015 Sep. PLoS Pathog. 2015. PMID: 26378441 Free PMC article. No abstract available.
Abstract
The immune system can recognize virtually any antigen, yet T cell responses against several pathogens, including Mycobacterium tuberculosis, are restricted to a limited number of immunodominant epitopes. The host factors that affect immunodominance are incompletely understood. Whether immunodominant epitopes elicit protective CD8+ T cell responses or instead act as decoys to subvert immunity and allow pathogens to establish chronic infection is unknown. Here we show that anatomically distinct human granulomas contain clonally expanded CD8+ T cells with overlapping T cell receptor (TCR) repertoires. Similarly, the murine CD8+ T cell response against M. tuberculosis is dominated by TB10.44-11-specific T cells with extreme TCRβ bias. Using a retro genic model of TB10.44-11-specific CD8+ Tcells, we show that TCR dominance can arise because of competition between clonotypes driven by differences in affinity. Finally, we demonstrate that TB10.4-specific CD8+ T cells mediate protection against tuberculosis, which requires interferon-γ production and TAP1-dependent antigen presentation in vivo. Our study of how immunodominance, biased TCR repertoires, and protection are inter-related, provides a new way to measure the quality of T cell immunity, which if applied to vaccine evaluation, could enhance our understanding of how to elicit protective T cell immunity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


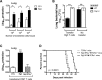
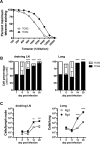
Similar articles
-
A Higher Activation Threshold of Memory CD8+ T Cells Has a Fitness Cost That Is Modified by TCR Affinity during Tuberculosis.PLoS Pathog. 2016 Jan 8;12(1):e1005380. doi: 10.1371/journal.ppat.1005380. eCollection 2016 Jan. PLoS Pathog. 2016. PMID: 26745507 Free PMC article.
-
Mycobacterium tuberculosis directs immunofocusing of CD8+ T cell responses despite vaccination.J Immunol. 2011 Feb 1;186(3):1627-37. doi: 10.4049/jimmunol.1002911. Epub 2010 Dec 22. J Immunol. 2011. PMID: 21178003 Free PMC article.
-
High frequency of CD4+ T cells specific for the TB10.4 protein correlates with protection against Mycobacterium tuberculosis infection.Infect Immun. 2006 Jun;74(6):3396-407. doi: 10.1128/IAI.02086-05. Infect Immun. 2006. PMID: 16714570 Free PMC article.
-
Antigen-specific CD8(+) T cells and protective immunity to tuberculosis.Adv Exp Med Biol. 2013;783:141-63. doi: 10.1007/978-1-4614-6111-1_8. Adv Exp Med Biol. 2013. PMID: 23468108 Free PMC article. Review.
-
Revealing factors determining immunodominant responses against dominant epitopes.Immunogenetics. 2020 Feb;72(1-2):109-118. doi: 10.1007/s00251-019-01134-9. Epub 2019 Dec 6. Immunogenetics. 2020. PMID: 31811313 Free PMC article. Review.
Cited by
-
A natural polymorphism of Mycobacterium tuberculosis in the esxH gene disrupts immunodomination by the TB10.4-specific CD8 T cell response.PLoS Pathog. 2020 Oct 19;16(10):e1009000. doi: 10.1371/journal.ppat.1009000. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33075106 Free PMC article.
-
Mucosal-associated invariant and γδ T cell subsets respond to initial Mycobacterium tuberculosis infection.JCI Insight. 2018 Oct 4;3(19):e121899. doi: 10.1172/jci.insight.121899. JCI Insight. 2018. PMID: 30282828 Free PMC article.
-
Mycobacterium tuberculosis-specific CD4+ and CD8+ T cells differ in their capacity to recognize infected macrophages.PLoS Pathog. 2018 May 21;14(5):e1007060. doi: 10.1371/journal.ppat.1007060. eCollection 2018 Feb. PLoS Pathog. 2018. PMID: 29782535 Free PMC article.
-
IL-21 signaling is essential for optimal host resistance against Mycobacterium tuberculosis infection.Sci Rep. 2016 Nov 7;6:36720. doi: 10.1038/srep36720. Sci Rep. 2016. PMID: 27819295 Free PMC article.
-
A Higher Activation Threshold of Memory CD8+ T Cells Has a Fitness Cost That Is Modified by TCR Affinity during Tuberculosis.PLoS Pathog. 2016 Jan 8;12(1):e1005380. doi: 10.1371/journal.ppat.1005380. eCollection 2016 Jan. PLoS Pathog. 2016. PMID: 26745507 Free PMC article.
References
-
- Davis MM, Bjorkman PJ (1988) T-cell antigen receptor genes and T-cell recognition [published erratum appears in Nature 1988 Oct 20;335(6192):744]. Nature 334: 395–402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

