Genomic analysis reveals the molecular basis for capsule loss in the group B Streptococcus population
- PMID: 25946017
- PMCID: PMC4422693
- DOI: 10.1371/journal.pone.0125985
Genomic analysis reveals the molecular basis for capsule loss in the group B Streptococcus population
Abstract
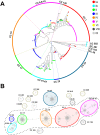
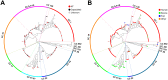
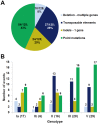
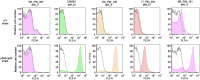
The human and bovine bacterial pathogen Streptococcus agalactiae (Group B Streptococcus, GBS) expresses a thick polysaccharide capsule that constitutes a major virulence factor and vaccine target. GBS can be classified into ten distinct serotypes differing in the chemical composition of their capsular polysaccharide. However, non-typeable strains that do not react with anti-capsular sera are frequently isolated from colonized and infected humans and cattle. To gain a comprehensive insight into the molecular basis for the loss of capsule expression in GBS, a collection of well-characterized non-typeable strains was investigated by genome sequencing. Genome based phylogenetic analysis extended to a wide population of sequenced strains confirmed the recently observed high clonality among GBS lineages mainly containing human strains, and revealed a much higher degree of diversity in the bovine population. Remarkably, non-typeable strains were equally distributed in all lineages. A number of distinct mutations in the cps operon were identified that were apparently responsible for inactivation of capsule synthesis. The most frequent genetic alterations were point mutations leading to stop codons in the cps genes, and the main target was found to be cpsE encoding the portal glycosyl transferase of capsule biosynthesis. Complementation of strains carrying missense mutations in cpsE with a wild-type gene restored capsule expression allowing the identification of amino acid residues essential for enzyme activity.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

