Multi-input distributed classifiers for synthetic genetic circuits
- PMID: 25946237
- PMCID: PMC4450813
- DOI: 10.1371/journal.pone.0125144
Multi-input distributed classifiers for synthetic genetic circuits
Abstract
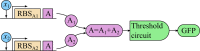
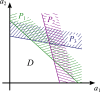
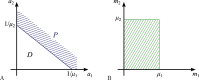
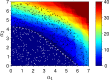
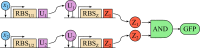
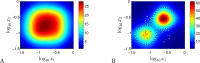
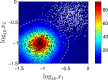
For practical construction of complex synthetic genetic networks able to perform elaborate functions it is important to have a pool of relatively simple modules with different functionality which can be compounded together. To complement engineering of very different existing synthetic genetic devices such as switches, oscillators or logical gates, we propose and develop here a design of synthetic multi-input classifier based on a recently introduced distributed classifier concept. A heterogeneous population of cells acts as a single classifier, whose output is obtained by summarizing the outputs of individual cells. The learning ability is achieved by pruning the population, instead of tuning parameters of an individual cell. The present paper is focused on evaluating two possible schemes of multi-input gene classifier circuits. We demonstrate their suitability for implementing a multi-input distributed classifier capable of separating data which are inseparable for single-input classifiers, and characterize performance of the classifiers by analytical and numerical results. The simpler scheme implements a linear classifier in a single cell and is targeted at separable classification problems with simple class borders. A hard learning strategy is used to train a distributed classifier by removing from the population any cell answering incorrectly to at least one training example. The other scheme implements a circuit with a bell-shaped response in a single cell to allow potentially arbitrary shape of the classification border in the input space of a distributed classifier. Inseparable classification problems are addressed using soft learning strategy, characterized by probabilistic decision to keep or discard a cell at each training iteration. We expect that our classifier design contributes to the development of robust and predictable synthetic biosensors, which have the potential to affect applications in a lot of fields, including that of medicine and industry.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

