Influence of in situ progressive N-terminal is still controversial truncation of glycogen branching enzyme in Escherichia coli DH5α on glycogen structure, accumulation, and bacterial viability
- PMID: 25947105
- PMCID: PMC4433092
- DOI: 10.1186/s12866-015-0421-9
Influence of in situ progressive N-terminal is still controversial truncation of glycogen branching enzyme in Escherichia coli DH5α on glycogen structure, accumulation, and bacterial viability
Abstract
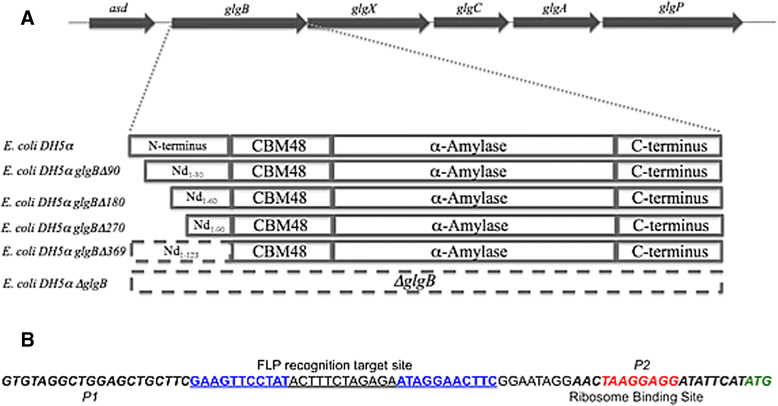
Background: Glycogen average chain length (ACL) has been linked with bacterial durability, but this was on the basis of observations across different species. We therefore wished to investigate the relationship between bacterial durability and glycogen ACL by varying glycogen average chain length in a single species. It has been shown that progressive shortening of the N-terminus of glycogen branching enzyme (GBE) leads to a lengthening of oligosaccharide inter-α-1,6-glycosidic chain lengths, so we sought to harness this to create a set of Escherichia coli DH5α strains with a range of glycogen average chain lengths, and assess these strains for durability related attributes, such as starvation, cold and desiccation stress resistance, and biofilm formation.
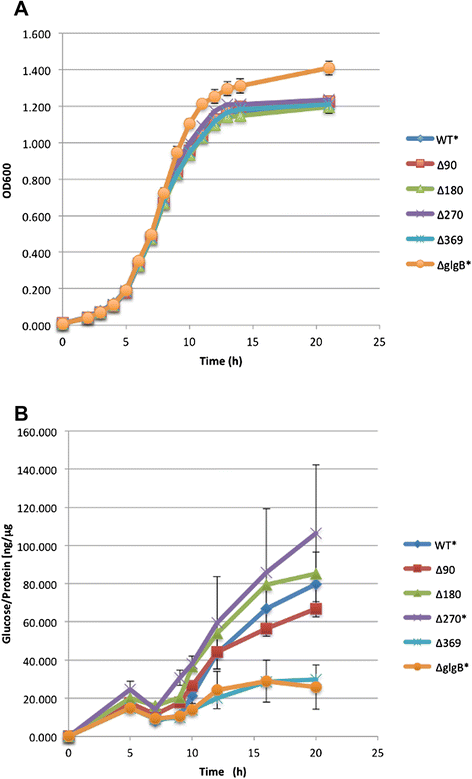
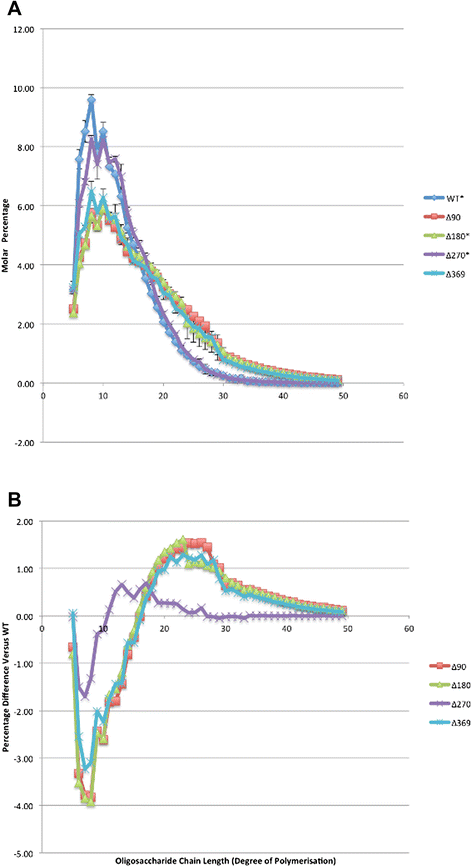
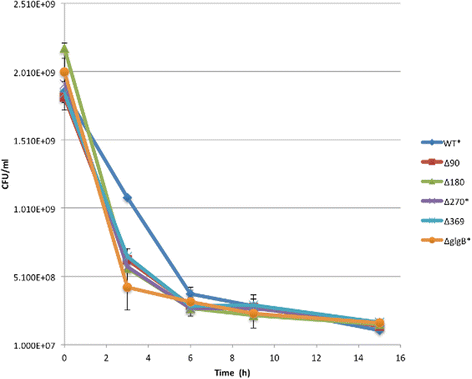
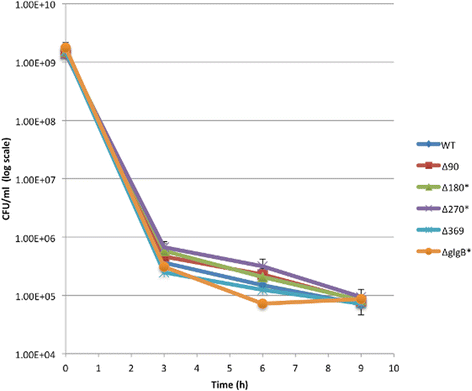
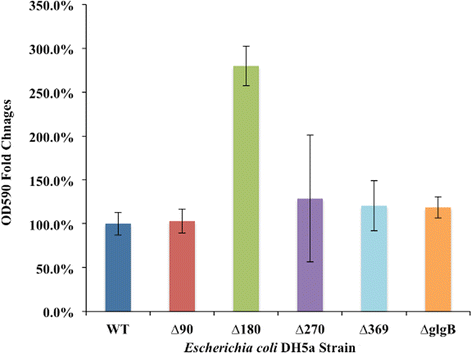
Results: A series of Escherichia coli DH5α mutants were created with glgB genes that were in situ progressively N-terminus truncated. N-terminal truncation shifted the distribution of glycogen chain lengths from 5-11 DP toward 13-50 DP, but the relationship between glgB length and glycogen ACL was not linear. Surprisingly, removal of the first 270 nucleotides of glgB (glgBΔ270) resulted in comparatively high glycogen accumulation, with the glycogen having short ACL. Complete knockout of glgB led to the formation of amylose-like glycogen containing long, linear α1,4-glucan chains with significantly reduced branching frequency. Physiologically, the set of mutant strains had reduced bacterial starvation resistance, while minimally increasing bacterial desiccation resistance. Finally, although there were no obvious changes in cold stress resistance or biofilm forming ability, one strain (glgBΔ180) had significantly increased biofilm formation in favourable media.
Conclusions: Despite glgB being the first gene of an operon, it is clear that in situ mutation is a viable means to create more biologically relevant mutant strains. Secondly, there was the suggestion in the data that impairments of starvation, cold and desiccation resistance were worse for the strain lacking glgB, though the first of these was not statistically significant. The results provide prima facie evidence linking abiotic stress tolerance with shorter glycogen ACL. However, further work needs to be done, perhaps in a less labile species. Further work is also required to tease out the complex relationship between glycogen abundance and glycogen structure.
Figures
References
-
- Preiss J. Glycogen Biosynthesis. In: Schaechter M, editor. Encyclopedia of Microbiology. 3. Oxford: Elsevier; 2009. pp. 145–158.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

