Atg1 family kinases in autophagy initiation
- PMID: 25948417
- PMCID: PMC4506457
- DOI: 10.1007/s00018-015-1917-z
Atg1 family kinases in autophagy initiation
Abstract
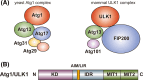
Autophagosome formation, a landmark event in autophagy, is accomplished by the concerted actions of Atg proteins. Among all Atg proteins, Atg1 kinase in yeast and its counterpart in higher eukaryotes, ULK1 kinase, function as the most upstream factor in this process and mediate autophagy initiation. In this review, we summarize current knowledge of the structure, molecular function, and regulation of Atg1 family kinases in the initiation of autophagy.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

