JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis
- PMID: 25948511
- PMCID: PMC4432656
- DOI: 10.1038/ncomms8052
JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis
Abstract
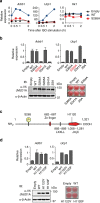
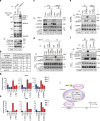
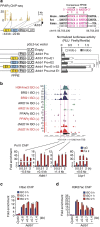
Histone 3 lysine 9 (H3K9) demethylase JMJD1A regulates β-adrenergic-induced systemic metabolism and body weight control. Here we show that JMJD1A is phosphorylated at S265 by protein kinase A (PKA), and this is pivotal to activate the β1-adrenergic receptor gene (Adrb1) and downstream targets including Ucp1 in brown adipocytes (BATs). Phosphorylation of JMJD1A by PKA increases its interaction with the SWI/SNF nucleosome remodelling complex and DNA-bound PPARγ. This complex confers β-adrenergic-induced rapid JMJD1A recruitment to target sites and facilitates long-range chromatin interactions and target gene activation. This rapid gene induction is dependent on S265 phosphorylation but not on demethylation activity. Our results show that JMJD1A has two important roles in regulating hormone-stimulated chromatin dynamics that modulate thermogenesis in BATs. In one role, JMJD1A is recruited to target sites and functions as a cAMP-responsive scaffold that facilitates long-range chromatin interactions, and in the second role, JMJD1A demethylates H3K9 di-methylation.
Figures





References
-
- Nedergaard J. et al.. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochim. Biophys. Acta 1504, 82–106 (2001). - PubMed
-
- Kooistra S. M. & Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13, 297–311 (2012). - PubMed
-
- Yamane K. et al.. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

