Disruption of the lamin A and matrin-3 interaction by myopathic LMNA mutations
- PMID: 25948554
- PMCID: PMC4492393
- DOI: 10.1093/hmg/ddv160
Disruption of the lamin A and matrin-3 interaction by myopathic LMNA mutations
Abstract
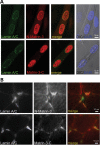
The nuclear face of the nuclear membrane is enriched with the intermediate filament protein lamin A. Mutations in LMNA, the gene encoding lamin A, lead to a diverse set of inherited conditions including myopathies that affect both the heart and skeletal muscle. To gain insight about lamin A protein interactions, binding proteins associated with the tail of lamin A were characterized. Of 130 nuclear proteins found associated with the lamin A tail, 17 (13%) were previously described lamin A binding partners. One protein not previously linked to lamin A, matrin-3, was selected for further study, because like LMNA mutations, matrin-3 has also been implicated in inherited myopathy. Matrin-3 binds RNA and DNA and is a nucleoplasmic protein originally identified from the insoluble nuclear fraction, referred to as the nuclear matrix. Anti-matrin-3 antibodies were found to co-immunoprecipitate lamin A, and the lamin-A binding domain was mapped to the carboxy-terminal half of matrin-3. Three-dimensional mapping of the lamin A-matrin-3 interface showed that the LMNA truncating mutation Δ303, which lacks the matrin-3 binding domain, was associated with an increased distance between lamin A and matrin-3. LMNA mutant cells are known to have altered biophysical properties and the matrin-3-lamin A interface is positioned to contribute to these defects.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures







References
-
- Gruenbaum Y., Margalit A., Goldman R.D., Shumaker D.K., Wilson K.L. (2005) The nuclear lamina comes of age. Nat. Rev. Mol. Cell. Biol., 6, 21–31. - PubMed
-
- Bertrand A.T., Chikhaoui K., Yaou R.B., Bonne G. (2011) Clinical and genetic heterogeneity in laminopathies. Biochem. Soc. Trans., 39, 1687–1692. - PubMed
-
- Krimm I., Ostlund C., Gilquin B., Couprie J., Hossenlopp P., Mornon J.P., Bonne G., Courvalin J.C., Worman H.J., Zinn-Justin S. (2002) The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure (Camb), 10, 811–823. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

