Fine Mapping Causal Variants with an Approximate Bayesian Method Using Marginal Test Statistics
- PMID: 25948564
- PMCID: PMC4512539
- DOI: 10.1534/genetics.115.176107
Fine Mapping Causal Variants with an Approximate Bayesian Method Using Marginal Test Statistics
Abstract
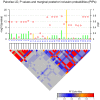
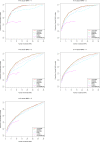
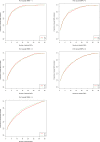
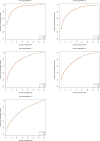
Two recently developed fine-mapping methods, CAVIAR and PAINTOR, demonstrate better performance over other fine-mapping methods. They also have the advantage of using only the marginal test statistics and the correlation among SNPs. Both methods leverage the fact that the marginal test statistics asymptotically follow a multivariate normal distribution and are likelihood based. However, their relationship with Bayesian fine mapping, such as BIMBAM, is not clear. In this study, we first show that CAVIAR and BIMBAM are actually approximately equivalent to each other. This leads to a fine-mapping method using marginal test statistics in the Bayesian framework, which we call CAVIAR Bayes factor (CAVIARBF). Another advantage of the Bayesian framework is that it can answer both association and fine-mapping questions. We also used simulations to compare CAVIARBF with other methods under different numbers of causal variants. The results showed that both CAVIARBF and BIMBAM have better performance than PAINTOR and other methods. Compared to BIMBAM, CAVIARBF has the advantage of using only marginal test statistics and takes about one-quarter to one-fifth of the running time. We applied different methods on two independent cohorts of the same phenotype. Results showed that CAVIARBF, BIMBAM, and PAINTOR selected the same top 3 SNPs; however, CAVIARBF and BIMBAM had better consistency in selecting the top 10 ranked SNPs between the two cohorts. Software is available at https://bitbucket.org/Wenan/caviarbf.
Keywords: Bayesian fine mapping; causal variants; marginal test statistics.
Copyright © 2015 by the Genetics Society of America.
Figures
References
-
- Armitage P., 1955. Tests for linear trends in proportions and frequencies. Biometrics 11: 375–386.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

