Identifying driver genes in cancer by triangulating gene expression, gene location, and survival data
- PMID: 25949096
- PMCID: PMC4354331
- DOI: 10.4137/CIN.S18302
Identifying driver genes in cancer by triangulating gene expression, gene location, and survival data
Abstract
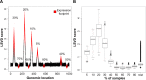
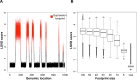
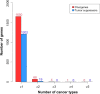
Driver genes are directly responsible for oncogenesis and identifying them is essential in order to fully understand the mechanisms of cancer. However, it is difficult to delineate them from the larger pool of genes that are deregulated in cancer (ie, passenger genes). In order to address this problem, we developed an approach called TRIAngulating Gene Expression (TRIAGE through clinico-genomic intersects). Here, we present a refinement of this approach incorporating a new scoring methodology to identify putative driver genes that are deregulated in cancer. TRIAGE triangulates - or integrates - three levels of information: gene expression, gene location, and patient survival. First, TRIAGE identifies regions of deregulated expression (ie, expression footprints) by deriving a newly established measure called the Local Singular Value Decomposition (LSVD) score for each locus. Driver genes are then distinguished from passenger genes using dual survival analyses. Incorporating measurements of gene expression and weighting them according to the LSVD weight of each tumor, these analyses are performed using the genes located in significant expression footprints. Here, we first use simulated data to characterize the newly established LSVD score. We then present the results of our application of this refined version of TRIAGE to gene expression data from five cancer types. This refined version of TRIAGE not only allowed us to identify known prominent driver genes, such as MMP1, IL8, and COL1A2, but it also led us to identify several novel ones. These results illustrate that TRIAGE complements existing tools, allows for the identification of genes that drive cancer and could perhaps elucidate potential future targets of novel anticancer therapeutics.
Keywords: cancer; data mining; driver genes; gene expression; survival.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

