Amyloid polymorphism: structural basis and neurobiological relevance
- PMID: 25950632
- PMCID: PMC4425266
- DOI: 10.1016/j.neuron.2015.03.017
Amyloid polymorphism: structural basis and neurobiological relevance
Abstract
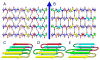
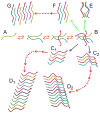
Our understanding of the molecular structures of amyloid fibrils that are associated with neurodegenerative diseases, of mechanisms by which disease-associated peptides and proteins aggregate into fibrils, and of structural properties of aggregation intermediates has advanced considerably in recent years. Detailed molecular structural models for certain fibrils and aggregation intermediates are now available. It is now well established that amyloid fibrils are generally polymorphic at the molecular level, with a given peptide or protein being capable of forming a variety of distinct, self-propagating fibril structures. Recent results from structural studies and from studies involving cell cultures, transgenic animals, and human tissue provide initial evidence that molecular structural variations in amyloid fibrils and related aggregates may correlate with or even produce variations in disease development. This article reviews our current knowledge of the structural and mechanistic aspects of amyloid formation, as well as current evidence for the biological relevance of structural variations.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

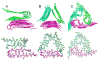
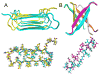
References
-
- Andronesi OC, von Bergen M, Biernat J, Seidel K, Griesinger C, Mandelkow E, Baldus M. Characterization of Alzheimer’s-like paired helical filaments from the core domain of tau protein using solid state NMR spectroscopy. J Am Chem Soc. 2008;130:5922–5928. - PubMed
-
- Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s β-amyloid fibrils from electron microscopy and solid state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

