Abnormal Localization and Tumor Suppressor Function of Epithelial Tissue-Specific Transcription Factor ESE3 in Esophageal Squamous Cell Carcinoma
- PMID: 25950810
- PMCID: PMC4423989
- DOI: 10.1371/journal.pone.0126319
Abnormal Localization and Tumor Suppressor Function of Epithelial Tissue-Specific Transcription Factor ESE3 in Esophageal Squamous Cell Carcinoma
Abstract
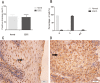
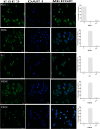
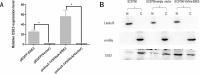
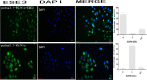
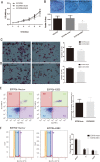
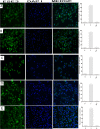
Esophageal cancer is one of the most common malignant cancers worldwide. The molecular mechanism of esophageal squamous cell carcinoma (ESCC) is still poorly understood. ESE3 is a member of the Ets transcription family, which is only expressed in epithelial tissues and acts as a tumor suppressor gene in prostate cancer. Our study aim was to confirm whether ESE3 is involved in the carcinogenesis of ESCC. Immunohistochemical analysis revealed that ESE3 was mainly located in cell nuclei of normal tissues and the cytoplasm in ESCC tissues. Immunofluorescence and western blot analyses of the normal esophageal cell line HEEpiC and ESCC cell lines EC9706 TE-1, KYSE150, and KYSE410 confirmed these results. pEGFP-ESE3 and pcDNA3.1-V5/HisA-ESE3 plasmids were constructed for overexpression of ESE3 in EC9706 and KYSE150 cells. The stably transfected cells showed restoration of the nuclear localization of ESE3. EC9706 cells with re-localization of ESE3 to the nucleus showed inhibition of proliferation, colony formation, migration, and invasion. To explore the possible mechanism of the differences in localization of ESE3 in normal esophageal cells and ESCC cells, ESCC cell lines were treated with the nuclear export inhibitor leptomycin B, transcription inhibitor actinomycin D, PKC inhibitor sphinganine, P38 MAPK inhibitor SB202190, and CK II inhibitor TBCA. These reagents were chosen according to the well-known mechanisms of protein translocation. However, the localization of ESE3 was unchanged after these treatments. The sequence of ESE3 cDNA in ESCC cells was identical to the standard sequence of ESE3 in the NCBI Genebank database, indicating that there was no mutation in the coding region of ESE3 in ESCC. Taken together, our study suggests that ESE3 plays an important role in the carcinogenesis of ESCC through changes in subcellular localization and may act as a tumor suppressor gene in ESCC, although the mechanisms require further study.
Conflict of interest statement
Figures


Similar articles
-
Epigenetic inactivation of secreted frizzled-related protein 2 in esophageal squamous cell carcinoma.World J Gastroenterol. 2012 Feb 14;18(6):532-40. doi: 10.3748/wjg.v18.i6.532. World J Gastroenterol. 2012. PMID: 22363119 Free PMC article.
-
Elevated maspin expression is associated with better overall survival in esophageal squamous cell carcinoma (ESCC).PLoS One. 2013 May 22;8(5):e63581. doi: 10.1371/journal.pone.0063581. Print 2013. PLoS One. 2013. PMID: 23717449 Free PMC article.
-
Profiling of differentially expressed cancer-related genes in esophageal squamous cell carcinoma (ESCC) using human cancer cDNA arrays: overexpression of oncogene MET correlates with tumor differentiation in ESCC.Clin Cancer Res. 2001 Nov;7(11):3519-25. Clin Cancer Res. 2001. PMID: 11705871
-
Targeted therapy of esophageal squamous cell carcinoma: the NRF2 signaling pathway as target.Ann N Y Acad Sci. 2018 Dec;1434(1):164-172. doi: 10.1111/nyas.13681. Epub 2018 May 11. Ann N Y Acad Sci. 2018. PMID: 29752726 Free PMC article. Review.
-
NOTCH and Esophageal Squamous Cell Carcinoma.Adv Exp Med Biol. 2021;1287:59-68. doi: 10.1007/978-3-030-55031-8_5. Adv Exp Med Biol. 2021. PMID: 33034026 Free PMC article. Review.
Cited by
-
ELF3, ELF5, EHF and SPDEF Transcription Factors in Tissue Homeostasis and Cancer.Molecules. 2018 Aug 30;23(9):2191. doi: 10.3390/molecules23092191. Molecules. 2018. PMID: 30200227 Free PMC article. Review.
-
Cpeb4-mediated Dclk2 promotes neuronal pyroptosis induced by chronic cerebral ischemia through phosphorylation of Ehf.J Cereb Blood Flow Metab. 2024 Sep;44(9):1655-1673. doi: 10.1177/0271678X241240590. Epub 2024 Mar 21. J Cereb Blood Flow Metab. 2024. PMID: 38513137 Free PMC article.
-
Comparative transcriptome characterization of esophageal squamous cell carcinoma and adenocarcinoma.Comput Struct Biotechnol J. 2023 Jul 25;21:3841-3853. doi: 10.1016/j.csbj.2023.07.030. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37564101 Free PMC article.
-
Increased expression of EHF via gene amplification contributes to the activation of HER family signaling and associates with poor survival in gastric cancer.Cell Death Dis. 2016 Oct 27;7(10):e2442. doi: 10.1038/cddis.2016.346. Cell Death Dis. 2016. PMID: 27787520 Free PMC article.
-
Increased expression of EHF contributes to thyroid tumorigenesis through transcriptionally regulating HER2 and HER3.Oncotarget. 2016 Sep 6;7(36):57978-57990. doi: 10.18632/oncotarget.11154. Oncotarget. 2016. PMID: 27517321 Free PMC article.
References
-
- Lehrbach DM, Nita ME, Cecconello I.Molecular aspects of esophageal squamous cell carcinoma carcinogenesis. Arq Gastroenterol. 2003; 40(4): 256–261. - PubMed
-
- Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004; 279(12): 11281–11292. - PubMed
-
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303(16): 11–34. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

