Marked Genetic Differentiation between Western Iberian and Italic Populations of the Olive Fly: Southern France as an Intermediate Area
- PMID: 25951107
- PMCID: PMC4423870
- DOI: 10.1371/journal.pone.0126702
Marked Genetic Differentiation between Western Iberian and Italic Populations of the Olive Fly: Southern France as an Intermediate Area
Abstract
The olive fly, Bactrocera oleae, is the most important pest affecting the olive industry, to which it is estimated to cause average annual losses in excess of one billion dollars. As with other insects with a wide distribution, it is generally accepted that the understanding of B. oleae population structure and dynamics is fundamental for the design and implementation of effective monitoring and control strategies. However, and despite important advances in the past decade, a clear picture of B. oleae's population structure is still lacking. In the Mediterranean basin, where more than 95% of olive production is concentrated, evidence from several studies suggests the existence of three distinct sub-populations, but the geographical limits of their distributions, and the level of interpenetration and gene flow among them remain ill-characterized. Here we use mitochondrial haplotype analysis to show that one of the Mediterranean mitochondrial lineages displays geographically correlated substructure and demonstrate that Italic populations, though markedly distinct from their Iberian and Levantine counterparts are more diverse than previously described. Finally, we show that this distinction does not result from extant hypothetical geographic limits imposed by the Alps or the Pyrenees nor, more generally, does it result from any sharp boundary, as intermixing is observed in a broad area, albeit at variable levels. Instead, Bayesian phylogeographic analysis suggests the interplay between isolation-mediated differentiation during glacial periods and bi-directional dispersal and population intermixing in the interglacials has played a major role in shaping current olive fly population structure.
Conflict of interest statement
Figures


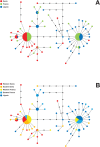

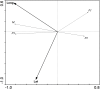

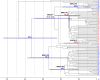
References
-
- Rice R, Phillips P, Stewart-Leslie J, Sibbett G. Olive fruit fly populations measured in Central and Southern California. Calif Agr 2003;57(4):122–7. 10.3733/ca.v057n04p122 - DOI
-
- Haniotakis GE. Olive pest control: present status and prospects. IOBC/WPRS Bulletins. 2005;28(9):1–9.
-
- Montiel Bueno A, Jones O. Alternative methods for controlling the olive fly, Bactrocera oleae, involving semiochemicals. IOBC/WPRS Bulletin. 2002;25(9):147–56.
-
- Segura MD, Callejas C, Ochando MD. Bactrocera oleae: a single large population in Northern Mediterranean basin. J Appl Ent. 2008;(132):706–13. 10.1111/j.1439-0418.2008.01366.x - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

