Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM
- PMID: 25953390
- PMCID: PMC4489599
- DOI: 10.1038/modpathol.2015.52
Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM
Abstract
High-risk human papillomavirus (HPV) types cause cervical lesions of varying severity, ranging from transient productive infections to high-grade neoplasia. Disease stratification requires the examination of lesional pathology, and possibly also the detection of biomarkers. P16(INK4a) and MCM are established surrogates of high-risk HPV E6/E7 activity, and can be extensively expressed in high-grade lesions. Here we have combined these two cellular biomarkers with detection of the abundant HPV-encoded E4 protein in order to identify both productive and transforming lesions. This approach has allowed us to distinguish true papillomavirus infections from similar pathologies, and has allowed us to divide the heterogeneous CIN2 category into those that are CIN1-like and express E4, and those that more closely resemble nonproductive CIN3. To achieve this, 530 lesional areas were evaluated according to standard pathology criteria and by using a multiple staining approach that allows us to superimpose biomarker patterns either singly or in combination onto an annotated hematoxylin and eosin (H&E) image. Conventional grading of neoplasia was established by review panel, and compared directly with the composite molecular pathology visualized on the same tissue section. The detection of E4 coincided with the onset of vacuolation, becoming abundant in koilocytes as the MCM marker declined and cells lost their defined nuclear margins as visualized by standard H&E staining. Of the dual marker approaches, p16(INK4a) and E4 appeared most promising, with E4 generally identifying areas of low-grade disease even when p16(INK4a) was present. Extensive p16(INK4a) expression usually coincided with an absence of E4 expression or its focal retention in sporadic cells within the lesion. Our results suggest that a straightforward molecular evaluation of HPV life-cycle deregulation in cervical neoplasia may help improve disease stratification, and that this can be achieved using complementary molecular biomarker pairs such as MCM/E4 or, more promisingly, p16(INK4a)/E4 as an adjunct to conventional pathology.
Figures
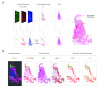
i) Most CIN1 are productive infections, with MCM (red) reaching and extending into the E4 expressing layers (green). In this lesion, MCM and p16INK4a have broadly similar distributions.
ii) Although p16INK4a (brown) and MCM (red) expression can have different distributions, MCM again extends into the E4-positive layers (green). In this lesion, MCM extends higher into the epithelial layers than p16INK4a.
iii) A small number of consensus CIN1 (6/33) failed to show E4 (green) biomarker expression. p16INK4a (brown) expression extends into the upper epithelial layers and is more extensive than MCM (red).
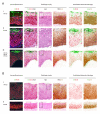
i) In the majority of CIN1, the E4 (green) biomarker accumulates during the process of koilocyte formation. Arrows labelled 1 to 4 show the progressive vacuolation that accompanies E4 accumulation (beginning at arrow 2 and highest at arrow 4), and the loss of the MCM biomarker (lowest at arrow 4).
ii) Despite some differences in cell morphology and tissue architecture amongst CIN1, the distribution of biomarkers described above (i) is conserved.
iii) In one (out of 33) consensus CIN1, koilocyte-like cells we present but were devoid of markers of productive infection, including the E4 (green) biomarker and associated MCM (red) staining.
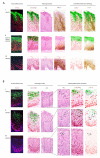
i) In the majority of CIN3, the E4 (green) biomarker is absent, and both the MCM (red) and p16INK4a (brown) biomarkers extend uniformly through the full thickness of the epithelium.
ii) Despite differences in lesion size and morphology, the biomarker patterns described above (i) were broadly similar in other CIN3.
i) In a small number of CIN3 (4/68), focal areas of E4 (green) were apparent. Both p16INK4a (brown) and MCM (red) extended through the full thickness of the lesion with evidence of nuclear crowding as seen in Fig 3A. Panel A is shown enlarged in Fig. 3C.
ii) In one case (out of 68), the E4 (green) biomarker was extensive, but was confined to regions showing low-grade pathology that lacked strong p16INK4a (brown) biomarker staining. Panel B is shown enlarged in Fig. 3B.
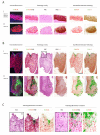
i) High cell density and the expression of MCM precedes the accumulation of E4 in a subset of cells showing evidence of vacuolation. P16INK4a and MCM expression extend throughout the full thickness of the epithelium. Panel A is enlarged in Fig.5.
ii) Despite differences in epithelial thickness, the biomarker pattern is preserved in other consensus CIN2.
iii) As in CIN1, some CIN2 show a more extensive expression of MCM into the upper epithelial layers when compared to p16INK4a. E4 expression is limited to cells close to the epithelial surface. Panel B is enlarged in Fig.5 to illustrate the correlation between pathology and biomarker patterns.
i) Loss of the E4 biomarker in a proportion of CIN2, can be associated with extensive expression of the p16INK4a /MCM biomarkers throughout the thickness of the epithelium, as well as an absence of obvious differentiation at the level of pathology. Panel C is enlarged in Fig. 5.
ii) MCM can extend closer throughout the epithelium more robustly than p16INK4a without the expression of the E4 biomarker. The panel shown in D is enlarged in Fig. 5 to show the absence of key pathology features apparent in the E4-positive CIN2.
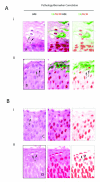
i) Pathology associated with E4 expression in CIN2. The six images show enlargements of the regions that are boxed in Fig. 4A(i) and (iii) (H&E images). The pattern of vacuolation and koilocyte formation is similar to that seen in CIN1 (Fig. 2B), with E4 expression, vacuolation and MCM decline coinciding closely (arrows 1&2).
ii) The six images show an enlargement of the regions that are boxed in Fig 4B(i) and (ii). In these lesions, vacuolated cells are sometimes apparent (arrows marked ‘v’), but are not necessarily associated with expression of the E4 biomarker (Fig. 4B(i) and (ii)).

Similar articles
-
Characterization of cervical biopsies of women with HIV and HPV co-infection using p16ink4a, ki-67 and HPV E4 immunohistochemistry and DNA methylation.Mod Pathol. 2020 Oct;33(10):1968-1978. doi: 10.1038/s41379-020-0528-x. Epub 2020 Apr 6. Mod Pathol. 2020. PMID: 32249820
-
Investigating Diagnostic Problems of CIN1 and CIN2 Associated With High-risk HPV by Combining the Novel Molecular Biomarker PanHPVE4 With P16INK4a.Am J Surg Pathol. 2015 Nov;39(11):1518-1528. doi: 10.1097/PAS.0000000000000498. Am J Surg Pathol. 2015. PMID: 26379150 Free PMC article.
-
Classification of high-grade cervical intraepithelial neoplasia by p16ink4a , Ki-67, HPV E4 and FAM19A4/miR124-2 methylation status demonstrates considerable heterogeneity with potential consequences for management.Int J Cancer. 2021 Aug 1;149(3):707-716. doi: 10.1002/ijc.33566. Epub 2021 May 11. Int J Cancer. 2021. PMID: 33729551 Free PMC article.
-
Efficiency of immunohistochemical p16 expression and HPV typing in cervical squamous intraepithelial lesion grading and review of the p16 literature.Pathol Res Pract. 2007;203(6):445-9. doi: 10.1016/j.prp.2007.03.010. Epub 2007 May 31. Pathol Res Pract. 2007. PMID: 17543474 Review.
-
Papillomavirus life cycle organization and biomarker selection.Dis Markers. 2007;23(4):297-313. doi: 10.1155/2007/613150. Dis Markers. 2007. PMID: 17627064 Free PMC article. Review.
Cited by
-
Expression of E4 Protein and HPV Major Capsid Protein (L1) as A Novel Combination in Squamous Intraepithelial Lesions.Biomedicines. 2023 Jan 16;11(1):225. doi: 10.3390/biomedicines11010225. Biomedicines. 2023. PMID: 36672733 Free PMC article.
-
HPV16 and 18 genome amplification show different E4-dependence, with 16E4 enhancing E1 nuclear accumulation and replicative efficiency via its cell cycle arrest and kinase activation functions.PLoS Pathog. 2017 Mar 17;13(3):e1006282. doi: 10.1371/journal.ppat.1006282. eCollection 2017 Mar. PLoS Pathog. 2017. PMID: 28306742 Free PMC article.
-
Expression of p16 and HPV E4 on biopsy samples and methylation of FAM19A4 and miR124-2 on cervical cytology samples in the classification of cervical squamous intraepithelial lesions.Cancer Med. 2020 Apr;9(7):2454-2461. doi: 10.1002/cam4.2855. Epub 2020 Feb 5. Cancer Med. 2020. PMID: 32022461 Free PMC article.
-
Three-tiered score for Ki-67 and p16ink4a improves accuracy and reproducibility of grading CIN lesions.J Clin Pathol. 2018 Nov;71(11):981-988. doi: 10.1136/jclinpath-2018-205271. Epub 2018 Jul 16. J Clin Pathol. 2018. PMID: 30012698 Free PMC article.
-
Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia.Viruses. 2015 Jul 16;7(7):3863-90. doi: 10.3390/v7072802. Viruses. 2015. PMID: 26193301 Free PMC article. Review.
References
-
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. - PubMed
-
- Doorbar J, Quint W, Banks L, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–70. - PubMed
-
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

