Challenges and opportunities of airborne metagenomics
- PMID: 25953766
- PMCID: PMC4453059
- DOI: 10.1093/gbe/evv064
Challenges and opportunities of airborne metagenomics
Abstract
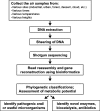
Recent metagenomic studies of environments, such as marine and soil, have significantly enhanced our understanding of the diverse microbial communities living in these habitats and their essential roles in sustaining vast ecosystems. The increase in the number of publications related to soil and marine metagenomics is in sharp contrast to those of air, yet airborne microbes are thought to have significant impacts on many aspects of our lives from their potential roles in atmospheric events such as cloud formation, precipitation, and atmospheric chemistry to their major impact on human health. In this review, we will discuss the current progress in airborne metagenomics, with a special focus on exploring the challenges and opportunities of undertaking such studies. The main challenges of conducting metagenomic studies of airborne microbes are as follows: 1) Low density of microorganisms in the air, 2) efficient retrieval of microorganisms from the air, 3) variability in airborne microbial community composition, 4) the lack of standardized protocols and methodologies, and 5) DNA sequencing and bioinformatics-related challenges. Overcoming these challenges could provide the groundwork for comprehensive analysis of airborne microbes and their potential impact on the atmosphere, global climate, and our health. Metagenomic studies offer a unique opportunity to examine viral and bacterial diversity in the air and monitor their spread locally or across the globe, including threats from pathogenic microorganisms. Airborne metagenomic studies could also lead to discoveries of novel genes and metabolic pathways relevant to meteorological and industrial applications, environmental bioremediation, and biogeochemical cycles.
Keywords: 16S rRNA sequencing; airborne microorganisms; culture-independent studies; metabolic potential; metagenomics; microbial diversity.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Amato P, et al. Microbial population in cloud water at the Puy de Dome: implications for the chemistry of clouds. Atmos Environ. 2005;39:4143–4153.
-
- Amato P, et al. A fate for organic acids, formaldehyde and methanol in cloud water: their biotransformation by micro-organisms. Atmos Chem Phys. 2007;7:4159–4169.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

