Identification of C2H2-ZF binding preferences from ChIP-seq data using RCADE
- PMID: 25953800
- PMCID: PMC4547615
- DOI: 10.1093/bioinformatics/btv284
Identification of C2H2-ZF binding preferences from ChIP-seq data using RCADE
Abstract
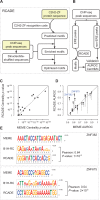
Current methods for motif discovery from chromatin immunoprecipitation followed by sequencing (ChIP-seq) data often identify non-targeted transcription factor (TF) motifs, and are even further limited when peak sequences are similar due to common ancestry rather than common binding factors. The latter aspect particularly affects a large number of proteins from the Cys2His2 zinc finger (C2H2-ZF) class of TFs, as their binding sites are often dominated by endogenous retroelements that have highly similar sequences. Here, we present recognition code-assisted discovery of regulatory elements (RCADE) for motif discovery from C2H2-ZF ChIP-seq data. RCADE combines predictions from a DNA recognition code of C2H2-ZFs with ChIP-seq data to identify models that represent the genuine DNA binding preferences of C2H2-ZF proteins. We show that RCADE is able to identify generalizable binding models even from peaks that are exclusively located within the repeat regions of the genome, where state-of-the-art motif finding approaches largely fail.
Availability and implementation: RCADE is available as a webserver and also for download at http://rcade.ccbr.utoronto.ca/.
Supplementary information: Supplementary data are available at Bioinformatics online.
Contact: t.hughes@utoronto.ca.
© The Author 2015. Published by Oxford University Press.
Figures
Similar articles
-
Computational Methods for Analysis of the DNA-Binding Preferences of Cys2His2 Zinc-Finger Proteins.Methods Mol Biol. 2018;1867:15-28. doi: 10.1007/978-1-4939-8799-3_2. Methods Mol Biol. 2018. PMID: 30155812
-
C2H2 zinc finger proteins greatly expand the human regulatory lexicon.Nat Biotechnol. 2015 May;33(5):555-62. doi: 10.1038/nbt.3128. Epub 2015 Feb 18. Nat Biotechnol. 2015. PMID: 25690854
-
Comparison of ChIP-Seq Data and a Reference Motif Set for Human KRAB C2H2 Zinc Finger Proteins.G3 (Bethesda). 2018 Jan 4;8(1):219-229. doi: 10.1534/g3.117.300296. G3 (Bethesda). 2018. PMID: 29146583 Free PMC article.
-
ChIP-Seq Data Analysis to Define Transcriptional Regulatory Networks.Adv Biochem Eng Biotechnol. 2017;160:1-14. doi: 10.1007/10_2016_43. Adv Biochem Eng Biotechnol. 2017. PMID: 28070596 Review.
-
An algorithmic perspective of de novo cis-regulatory motif finding based on ChIP-seq data.Brief Bioinform. 2018 Sep 28;19(5):1069-1081. doi: 10.1093/bib/bbx026. Brief Bioinform. 2018. PMID: 28334268 Review.
Cited by
-
Finding motifs using DNA images derived from sparse representations.Bioinformatics. 2023 Jun 1;39(6):btad378. doi: 10.1093/bioinformatics/btad378. Bioinformatics. 2023. PMID: 37294804 Free PMC article.
-
Transposable elements are dynamically regulated in medium spiny neurons and may contribute to the molecular and behavioral adaptations to cocaine.Biol Psychiatry. 2025 Jul 25:S0006-3223(25)01352-6. doi: 10.1016/j.biopsych.2025.07.014. Online ahead of print. Biol Psychiatry. 2025. PMID: 40716507
-
Young KRAB-zinc finger gene clusters are highly dynamic incubators of ERV-driven genetic heterogeneity in mice.bioRxiv [Preprint]. 2025 Mar 2:2025.02.26.640358. doi: 10.1101/2025.02.26.640358. bioRxiv. 2025. PMID: 40161592 Free PMC article. Preprint.
-
Low-Affinity Binding Sites and the Transcription Factor Specificity Paradox in Eukaryotes.Annu Rev Cell Dev Biol. 2019 Oct 6;35:357-379. doi: 10.1146/annurev-cellbio-100617-062719. Epub 2019 Jul 5. Annu Rev Cell Dev Biol. 2019. PMID: 31283382 Free PMC article. Review.
-
Extensive binding of uncharacterized human transcription factors to genomic dark matter.bioRxiv [Preprint]. 2024 Nov 12:2024.11.11.622123. doi: 10.1101/2024.11.11.622123. bioRxiv. 2024. PMID: 39605320 Free PMC article. Preprint.
References
-
- Bailey T.L., Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol., 2, 28–36. - PubMed
-
- Najafabadi H.S., et al. (2015) C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol., 33, 555–562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

