RAX2: a genome-wide detection method of condition-associated transcription variation
- PMID: 25953852
- PMCID: PMC4551904
- DOI: 10.1093/nar/gkv411
RAX2: a genome-wide detection method of condition-associated transcription variation
Abstract
Most mammalian genes have mRNA variants due to alternative promoter usage, alternative splicing, and alternative cleavage and polyadenylation. Expression of alternative RNA isoforms has been found to be associated with tumorigenesis, proliferation and differentiation. Detection of condition-associated transcription variation requires association methods. Traditional association methods such as Pearson chi-square test and Fisher Exact test are single test methods and do not work on count data with replicates. Although the Cochran Mantel Haenszel (CMH) approach can handle replicated count data, our simulations showed that multiple CMH tests still had very low power. To identify condition-associated variation of transcription, we here proposed a ranking analysis of chi-squares (RAX2) for large-scale association analysis. RAX2 is a nonparametric method and has accurate and conservative estimation of FDR profile. Simulations demonstrated that RAX2 performs well in finding condition-associated transcription variants. We applied RAX2 to primary T-cell transcriptomic data and identified 1610 (16.3%) tags associated in transcription with immune stimulation at FDR < 0.05. Most of these tags also had differential expression. Analysis of two and three tags within genes revealed that under immune stimulation short RNA isoforms were preferably used.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
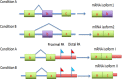
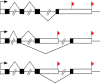
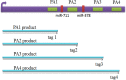
 ), four poly(A) sites (PA1, PA2, PA3 and PA4) in 3′ UTR coding for several mRNA isoforms that are different in their 3′ end. Alternative polyadenylation changes the length of the 3′ UTR and it also can change microRNA binding sites in 3′ UTR. While microRNAs tend to repress translation and promote degradation of the mRNAs they bind to. Since transcription products are one-to-one corresponding to poly(A) sites, we define the transcription products as tags.
), four poly(A) sites (PA1, PA2, PA3 and PA4) in 3′ UTR coding for several mRNA isoforms that are different in their 3′ end. Alternative polyadenylation changes the length of the 3′ UTR and it also can change microRNA binding sites in 3′ UTR. While microRNAs tend to repress translation and promote degradation of the mRNAs they bind to. Since transcription products are one-to-one corresponding to poly(A) sites, we define the transcription products as tags.



References
-
- Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

