Contrasting genetic architectures in different mouse reference populations used for studying complex traits
- PMID: 25953951
- PMCID: PMC4448675
- DOI: 10.1101/gr.187450.114
Contrasting genetic architectures in different mouse reference populations used for studying complex traits
Abstract
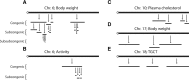
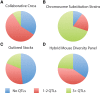
Quantitative trait loci (QTLs) are being used to study genetic networks, protein functions, and systems properties that underlie phenotypic variation and disease risk in humans, model organisms, agricultural species, and natural populations. The challenges are many, beginning with the seemingly simple tasks of mapping QTLs and identifying their underlying genetic determinants. Various specialized resources have been developed to study complex traits in many model organisms. In the mouse, remarkably different pictures of genetic architectures are emerging. Chromosome Substitution Strains (CSSs) reveal many QTLs, large phenotypic effects, pervasive epistasis, and readily identified genetic variants. In contrast, other resources as well as genome-wide association studies (GWAS) in humans and other species reveal genetic architectures dominated with a relatively modest number of QTLs that have small individual and combined phenotypic effects. These contrasting architectures are the result of intrinsic differences in the study designs underlying different resources. The CSSs examine context-dependent phenotypic effects independently among individual genotypes, whereas with GWAS and other mouse resources, the average effect of each QTL is assessed among many individuals with heterogeneous genetic backgrounds. We argue that variation of genetic architectures among individuals is as important as population averages. Each of these important resources has particular merits and specific applications for these individual and population perspectives. Collectively, these resources together with high-throughput genotyping, sequencing and genetic engineering technologies, and information repositories highlight the power of the mouse for genetic, functional, and systems studies of complex traits and disease models.
© 2015 Buchner and Nadeau; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Ackerman KG, Huang H, Grasemann H, Puma C, Singer JB, Hill AE, Lander E, Nadeau JH, Churchill GA, Drazen JM, et al. 2005. Interacting genetic loci cause airway hyperresponsiveness. Physiol Genomics 21: 105–111. - PubMed
-
- Alonso-Blanco C, Méndez-Vigo B. 2014. Genetic architecture of naturally occurring quantitative traits in plants: an updated synthesis. Curr Opin Plant Biol 18: 37–43. - PubMed
-
- Alves MM, Sribudiani Y, Brouwer RWW, Amiel J, Antiñolo G, Borrego S, Ceccherini I, Chakravarti A, Fernández RM, Garcia-Barcelo MM, et al. 2013. Contribution of rare and common variants determine complex diseases—Hirschsprung disease as a model. Dev Biol 382: 320–329. - PubMed
Publication types
MeSH terms
Grants and funding
- AR044927/AR/NIAMS NIH HHS/United States
- DK099533/DK/NIDDK NIH HHS/United States
- R01 CA075056/CA/NCI NIH HHS/United States
- DP1 HD075624/HD/NICHD NIH HHS/United States
- K01 DK084079/DK/NIDDK NIH HHS/United States
- R56 AR044927/AR/NIAMS NIH HHS/United States
- R01 AR044927/AR/NIAMS NIH HHS/United States
- CA75056/CA/NCI NIH HHS/United States
- DP1HD075624/DP/NCCDPHP CDC HHS/United States
- DA033646/DA/NIDA NIH HHS/United States
- R01 DA033646/DA/NIDA NIH HHS/United States
- R03 DK099533/DK/NIDDK NIH HHS/United States
- P40 RR012305/RR/NCRR NIH HHS/United States
- RR12305/RR/NCRR NIH HHS/United States
- DK084079/DK/NIDDK NIH HHS/United States
- R29 AR044927/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
