The landscape of genomic imprinting across diverse adult human tissues
- PMID: 25953952
- PMCID: PMC4484390
- DOI: 10.1101/gr.192278.115
The landscape of genomic imprinting across diverse adult human tissues
Abstract
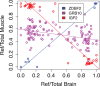
Genomic imprinting is an important regulatory mechanism that silences one of the parental copies of a gene. To systematically characterize this phenomenon, we analyze tissue specificity of imprinting from allelic expression data in 1582 primary tissue samples from 178 individuals from the Genotype-Tissue Expression (GTEx) project. We characterize imprinting in 42 genes, including both novel and previously identified genes. Tissue specificity of imprinting is widespread, and gender-specific effects are revealed in a small number of genes in muscle with stronger imprinting in males. IGF2 shows maternal expression in the brain instead of the canonical paternal expression elsewhere. Imprinting appears to have only a subtle impact on tissue-specific expression levels, with genes lacking a systematic expression difference between tissues with imprinted and biallelic expression. In summary, our systematic characterization of imprinting in adult tissues highlights variation in imprinting between genes, individuals, and tissues.
© 2015 Baran et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP. 2008. Global survey of genomic imprinting by transcriptome sequencing. Curr Biol 18: 1735–1741. - PubMed
Publication types
MeSH terms
Grants and funding
- 1K25HL121295-01A1/HL/NHLBI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- T32 GM067547/GM/NIGMS NIH HHS/United States
- 1R01GM110251-01/GM/NIGMS NIH HHS/United States
- DA006227/DA/NIDA NIH HHS/United States
- T32 EB009383/EB/NIBIB NIH HHS/United States
- R01 MH090936/MH/NIMH NIH HHS/United States
- MH090948/MH/NIMH NIH HHS/United States
- MH090936/MH/NIMH NIH HHS/United States
- HHSN26820100029C/PHS HHS/United States
- R01 MH090951/MH/NIMH NIH HHS/United States
- R01MH101814/MH/NIMH NIH HHS/United States
- K25 HL121295/HL/NHLBI NIH HHS/United States
- R01 DA006227/DA/NIDA NIH HHS/United States
- MH090937/MH/NIMH NIH HHS/United States
- R01 MH090948/MH/NIMH NIH HHS/United States
- R01 MH090941/MH/NIMH NIH HHS/United States
- MH090951/MH/NIMH NIH HHS/United States
- 1R01HL124285-01/HL/NHLBI NIH HHS/United States
- R01 GM110251/GM/NIGMS NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- R01 MH090937/MH/NIMH NIH HHS/United States
- HHSN268201000029C/HL/NHLBI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- R01 MH101814/MH/NIMH NIH HHS/United States
- R01 HL124285/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
