Human genomics. The human transcriptome across tissues and individuals
- PMID: 25954002
- PMCID: PMC4547472
- DOI: 10.1126/science.aaa0355
Human genomics. The human transcriptome across tissues and individuals
Abstract
Transcriptional regulation and posttranscriptional processing underlie many cellular and organismal phenotypes. We used RNA sequence data generated by Genotype-Tissue Expression (GTEx) project to investigate the patterns of transcriptome variation across individuals and tissues. Tissues exhibit characteristic transcriptional signatures that show stability in postmortem samples. These signatures are dominated by a relatively small number of genes—which is most clearly seen in blood—though few are exclusive to a particular tissue and vary more across tissues than individuals. Genes exhibiting high interindividual expression variation include disease candidates associated with sex, ethnicity, and age. Primary transcription is the major driver of cellular specificity, with splicing playing mostly a complementary role; except for the brain, which exhibits a more divergent splicing program. Variation in splicing, despite its stochasticity, may play in contrast a comparatively greater role in defining individual phenotypes.
Copyright © 2015, American Association for the Advancement of Science.
Figures

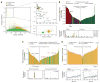
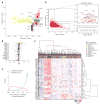
Comment in
-
Human genetics. GTEx detects genetic effects.Science. 2015 May 8;348(6235):640-1. doi: 10.1126/science.aab3002. Science. 2015. PMID: 25953996 No abstract available.
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

