Genome-wide comparison of Asian and African rice reveals high recent activity of DNA transposons
- PMID: 25954322
- PMCID: PMC4423477
- DOI: 10.1186/s13100-015-0040-x
Genome-wide comparison of Asian and African rice reveals high recent activity of DNA transposons
Abstract
Background: DNA (Class II) transposons are ubiquitous in plant genomes. However, unlike for (Class I) retrotransposons, only little is known about their proliferation mechanisms, activity, and impact on genomes. Asian and African rice (Oryza sativa and O. glaberrima) diverged approximately 600,000 years ago. Their fully sequenced genomes therefore provide an excellent opportunity to study polymorphisms introduced from recent transposon activity.
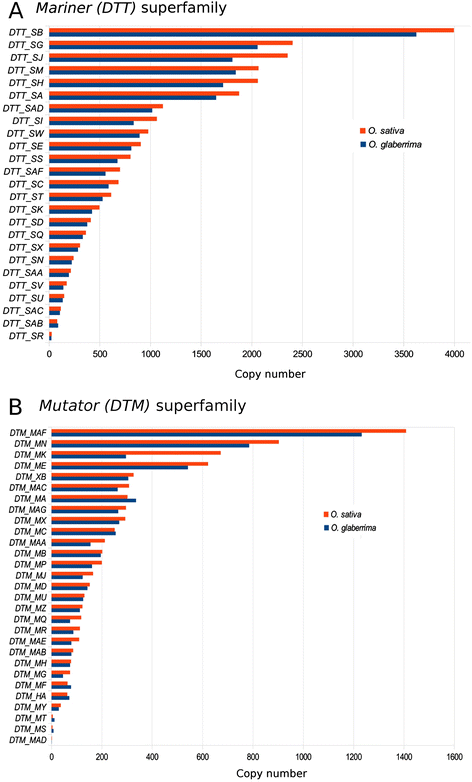
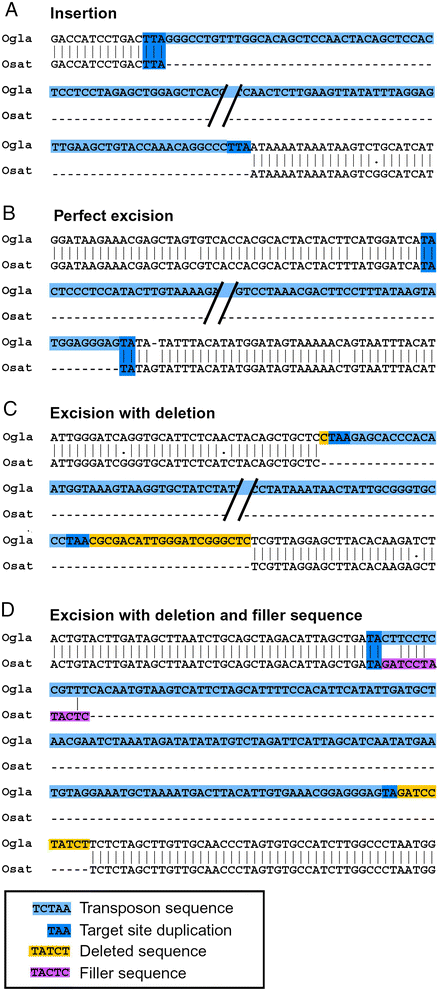
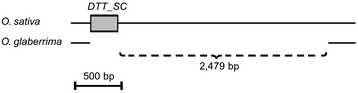
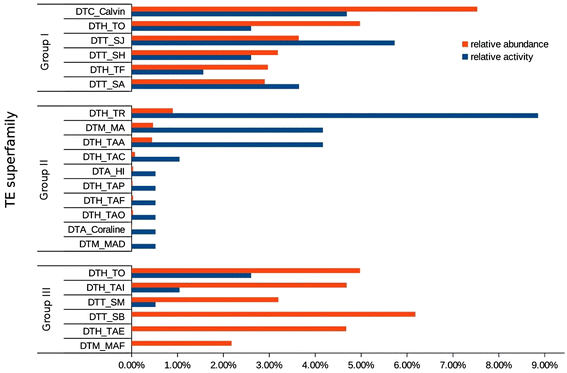
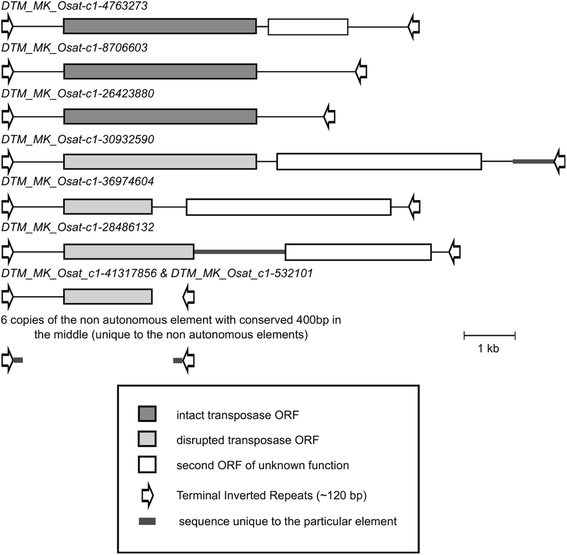
Results: We manually analyzed 1,821 transposon related polymorphisms among which we identified 487 loci which clearly resulted from DNA transposon insertions and excisions. In total, we estimate about 4,000 (3.5% of all DNA transposons) to be polymorphic between the two species, indicating a high level of transposable element (TE) activity. The vast majority of the recently active elements are non-autonomous. Nevertheless, we identified multiple potentially functional autonomous elements. Furthermore, we quantified the impacts of insertions and excisions on the adjacent sequences. Transposon insertions were found to be generally precise, creating simple target site duplications. In contrast, excisions almost always go along with the deletion of flanking sequences and/or the insertion of foreign 'filler' segments. Some of the excision-triggered deletions ranged from hundreds to thousands of bp flanking the excision site. Furthermore, we found in some superfamilies unexpectedly low numbers of excisions. This suggests that some excisions might cause such large-scale rearrangements so that they cannot be detected anymore.
Conclusions: We conclude that the activity of DNA transposons (particularly the excision process) is a major evolutionary force driving the generation of genetic diversity.
Keywords: DNA transposon activity; Proliferation mechanism; Rice.
Figures
Similar articles
-
DNA transposon activity is associated with increased mutation rates in genes of rice and other grasses.Nat Commun. 2016 Sep 7;7:12790. doi: 10.1038/ncomms12790. Nat Commun. 2016. PMID: 27599761 Free PMC article.
-
Inter-species sequence comparison of Brachypodium reveals how transposon activity corrodes genome colinearity.Plant J. 2012 Aug;71(4):550-63. doi: 10.1111/j.1365-313X.2012.05007.x. Epub 2012 Jun 18. Plant J. 2012. PMID: 22448600
-
Transposons play an important role in the evolution and diversification of centromeres among closely related species.Front Plant Sci. 2015 Apr 7;6:216. doi: 10.3389/fpls.2015.00216. eCollection 2015. Front Plant Sci. 2015. PMID: 25904926 Free PMC article.
-
Introduction of Plant Transposon Annotation for Beginners.Biology (Basel). 2023 Nov 26;12(12):1468. doi: 10.3390/biology12121468. Biology (Basel). 2023. PMID: 38132293 Free PMC article. Review.
-
Genome juggling by transposons: Tam3-induced rearrangements in Antirrhinum majus.Dev Genet. 1989;10(6):438-51. doi: 10.1002/dvg.1020100605. Dev Genet. 1989. PMID: 2557989 Review.
Cited by
-
The repetitive landscape of the 5100 Mbp barley genome.Mob DNA. 2017 Dec 20;8:22. doi: 10.1186/s13100-017-0102-3. eCollection 2017. Mob DNA. 2017. PMID: 29270235 Free PMC article.
-
DNA transposon activity is associated with increased mutation rates in genes of rice and other grasses.Nat Commun. 2016 Sep 7;7:12790. doi: 10.1038/ncomms12790. Nat Commun. 2016. PMID: 27599761 Free PMC article.
-
DNA Transposition at Work.Chem Rev. 2016 Oct 26;116(20):12758-12784. doi: 10.1021/acs.chemrev.6b00003. Epub 2016 May 17. Chem Rev. 2016. PMID: 27187082 Free PMC article. Review.
-
African Cultivated, Wild and Weedy Rice (Oryza spp.): Anticipating Further Genomic Studies.Biology (Basel). 2024 Sep 5;13(9):697. doi: 10.3390/biology13090697. Biology (Basel). 2024. PMID: 39336124 Free PMC article. Review.
-
The making of a genomic parasite - the Mothra family sheds light on the evolution of Helitrons in plants.Mob DNA. 2015 Dec 17;6:23. doi: 10.1186/s13100-015-0054-4. eCollection 2015. Mob DNA. 2015. PMID: 26688693 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

