In vitro stability of therapeutically relevant, internally truncated dystrophins
- PMID: 25954502
- PMCID: PMC4424174
- DOI: 10.1186/s13395-015-0040-z
In vitro stability of therapeutically relevant, internally truncated dystrophins
Abstract
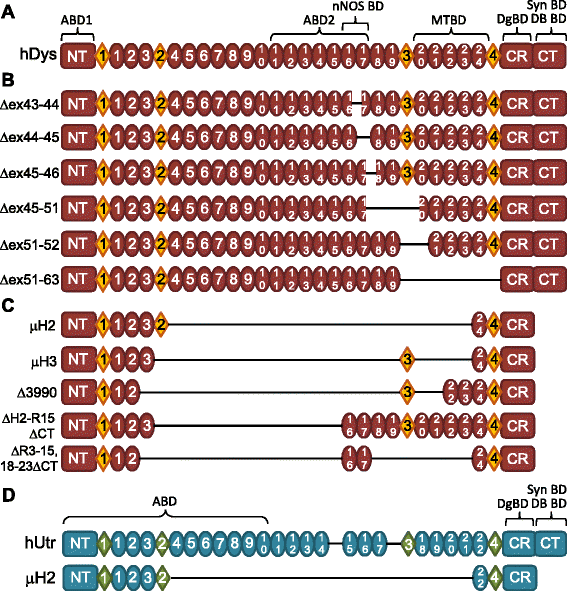
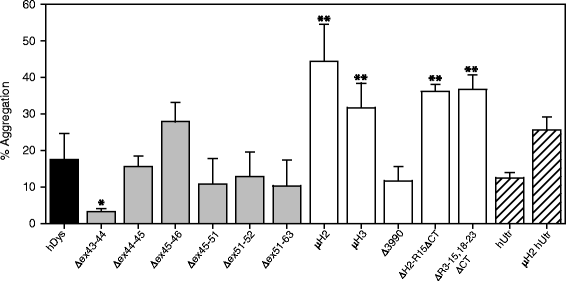
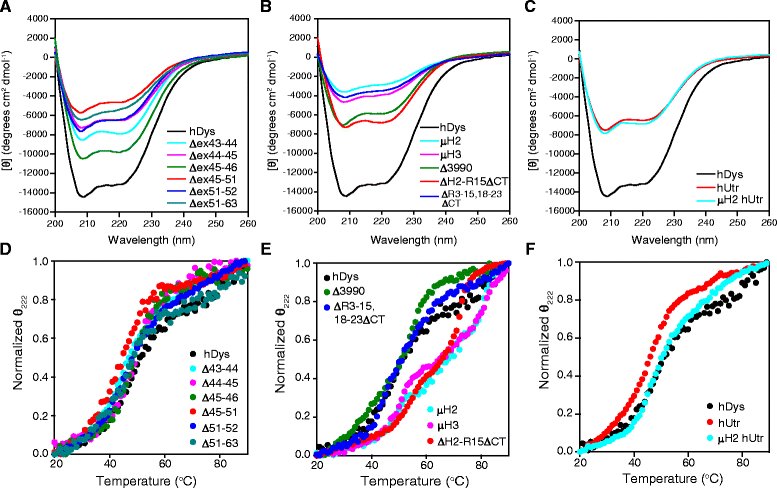
Background: The X-linked recessive disease Duchenne muscular dystrophy (DMD) is caused by mutations in the gene encoding the protein dystrophin. Despite its large size, dystrophin is a highly stable protein, demonstrating cooperative unfolding during thermal denaturation as monitored by circular dichroism spectroscopy. In contrast, internal sequence deletions have been associated with a loss of the cooperative unfolding and cause in vitro protein aggregation. Several emerging therapy options for DMD utilize internally deleted micro-dystrophins and multi-exon-skipped dystrophins that produce partially functional proteins, but the stability of such internally truncated proteins has not been investigated.
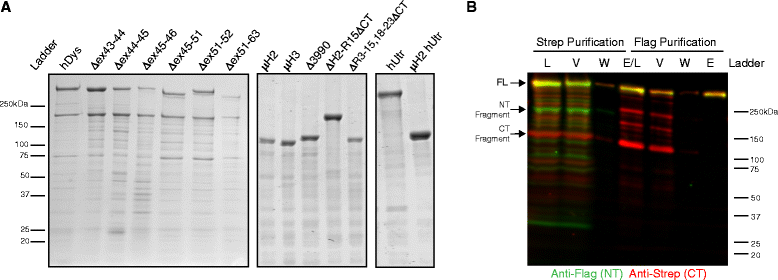
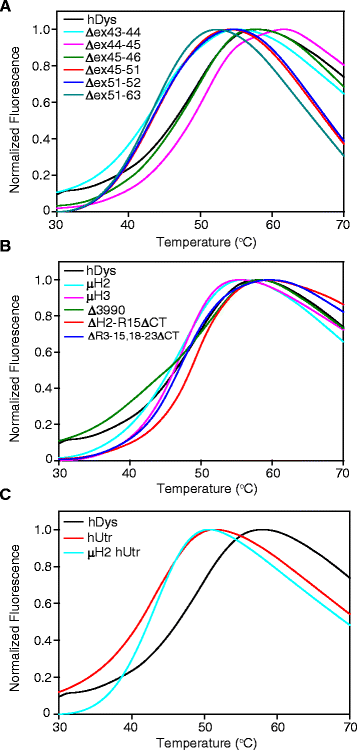
Methods: In this study, we analyzed the in vitro stability of human dystrophin constructs skipped around exon 45 or exon 51, several dystrophin gene therapy constructs, as well as human full-length and micro-utrophin. Constructs were expressed in insect cells using the baculovirus system, purified by affinity chromatography, and analyzed by high-speed sedimentation, circular dichroism spectroscopy, and differential scanning fluorimetry.
Results: Our results reveal that not all gene therapy constructs display stabilities consistent with full-length human dystrophin. However, all dystrophins skipped in-frame around exon 45 or exon 51 show stability profiles congruent with intact human dystrophin. Similar to previous studies of mouse proteins, full-length human utrophin also displays stability similar to human dystrophin and does not appear to be affected by a large internal deletion.
Conclusions: Our results suggest that the in vitro stability of human dystrophin is less sensitive to smaller deletions at natural exon boundaries than larger, more complex deletions present in some gene therapy constructs.
Keywords: Becker muscular dystrophy; Duchenne muscular dystrophy; Dystrophin; Exon skipping; Gene therapy; Utrophin.
Figures
References
-
- Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772:108–17 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

