Flanking A·T basepairs destabilize the B(∗) conformation of DNA A-tracts
- PMID: 25954886
- PMCID: PMC4423069
- DOI: 10.1016/j.bpj.2015.01.044
Flanking A·T basepairs destabilize the B(∗) conformation of DNA A-tracts
Abstract
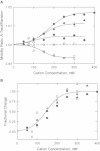
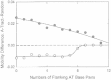
Capillary electrophoresis has been used to characterize the interaction of monovalent cations with 26-basepair DNA oligomers containing A-tracts embedded in flanking sequences with different basepair compositions. A 26-basepair random-sequence oligomer was used as the reference; lithium and tetrabutylammonium (TBA(+)) ions were used as the probe ions. The free solution mobilities of the A-tract and random-sequence oligomers were identical in solutions containing <∼ 100 mM cation. At higher cation concentrations, the A-tract oligomers migrated faster than the reference oligomer in TBA(+) and slower than the reference in Li(+). Hence, cations of different sizes can interact very differently with DNA A-tracts. The increased mobilities observed in TBA(+) suggest that the large hydrophobic TBA(+) ions are preferentially excluded from the vicinity of the A-tract minor groove, increasing the effective net charge of the A-tract oligomers and increasing the mobility. By contrast, Li(+) ions decrease the mobility of A-tract oligomers because of the preferential localization of Li(+) ions in the narrow A-tract minor groove. Embedding the A-tracts in AT-rich flanking sequences markedly alters preferential interactions of monovalent cations with the B(∗) conformation. Hence, A-tracts embedded in genomic DNA may or may not interact preferentially with monovalent cations, depending on the relative number of A · T basepairs in the flanking sequences.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Nelson H.C.M., Finch J.T., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. - PubMed
-
- Hagerman P.J. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 1990;59:755–781. - PubMed
-
- Olson W.K., Zhurkin V.B. Twenty years of DNA bending. In: Sarma R.H., Sarma M.H., editors. Biological Structure and Dynamics. Adenine Press; Schenectady, NY: 1996. pp. 341–370.
-
- Haran T.E., Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 2009;42:41–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

