Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores
- PMID: 25954891
- PMCID: PMC4423055
- DOI: 10.1016/j.bpj.2015.03.025
Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores
Abstract
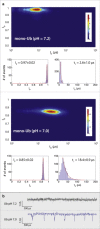
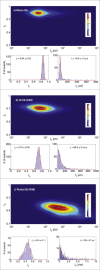
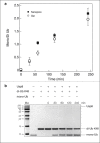
Nanopore sensing involves an electrophoretic transport of analytes through a nanoscale pore, permitting label-free sensing at the single-molecule level. However, to date, the detection of individual small proteins has been challenging, primarily due to the poor signal/noise ratio that these molecules produce during passage through the pore. Here, we show that fine adjustment of the buffer pH, close to the isoelectric point, can be used to slow down the translocation speed of the analytes, hence permitting sensing and characterization of small globular proteins. Ubiquitin (Ub) is a small protein of 8.5 kDa, which is well conserved in all eukaryotes. Ub conjugates to proteins as a posttranslational modification called ubiquitination. The immense diversity of Ub substrates, as well as the complexity of Ub modification types and the numerous physiological consequences of these modifications, make Ub and Ub chains an interesting and challenging subject of study. The ability to detect Ub and to identify Ub linkage type at the single-molecule level may provide a novel tool for investigation in the Ub field. This is especially adequate because, for most ubiquitinated substrates, Ub modifies only a few molecules in the cell at a given time. Applying our method to the detection of mono- and poly-Ub molecules, we show that we can analyze their characteristics using nanopores. Of particular importance is that two Ub dimers that are equal in molecular weight but differ in 3D structure due to their different linkage types can be readily discriminated. Thus, to our knowledge, our method offers a novel approach for analyzing proteins in unprecedented detail using solid-state nanopores. Specifically, it provides the basis for development of single-molecule sensing of differently ubiquitinated substrates with different biological significance. Finally, our study serves as a proof of concept for approaching nanopore detection of sub-10-kDa proteins and demonstrates the ability of this method to differentiate among native and untethered proteins of the same mass.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Vijay-Kumar S., Bugg C.E., Cook W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–544. - PubMed
-
- Varshavsky A. The ubiquitin system. Trends Biochem. Sci. 1997;22:383–387. - PubMed
-
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. - PubMed
-
- Komander D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009;37:937–953. - PubMed
-
- Komander D., Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

