Phylum level change in the cecal and fecal gut communities of rats fed diets containing different fermentable substrates supports a role for nitrogen as a factor contributing to community structure
- PMID: 25954902
- PMCID: PMC4446752
- DOI: 10.3390/nu7053279
Phylum level change in the cecal and fecal gut communities of rats fed diets containing different fermentable substrates supports a role for nitrogen as a factor contributing to community structure
Abstract
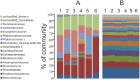
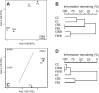
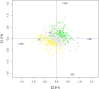
Fermentation differs between the proximal and distal gut but little is known regarding how the bacterial communities differ or how they are influenced by diet. In order to investigate this, we compared community diversity in the cecum and feces of rats by 16S rRNA gene content and DNA shot gun metagenomics after feeding purified diets containing different fermentable substrates. Gut community composition was dependent on the source of fermentable substrate included in the diet. Cecal communities were dominated by Firmicutes, and contained a higher abundance of Lachnospiraceae compared to feces. In feces, community structure was shifted by varying degrees depending on diet towards the Bacteroidetes, although this change was not always evident from 16S rRNA gene data. Multi-dimensional scaling analysis (PCoA) comparing cecal and fecal metagenomes grouped by location within the gut rather than by diet, suggesting that factors in addition to substrate were important for community change in the distal gut. Differentially abundant genes in each environment supported this shift away from the Firmicutes in the cecum (e.g., motility) towards the Bacteroidetes in feces (e.g., Bacteroidales transposons). We suggest that this phylum level change reflects a shift to ammonia as the primary source of nitrogen used to support continued microbial growth in the distal gut.
Figures

References
-
- Swanson K.S., Dowd S.E., Suchodolski J.S., Middelbos I.S., Vester B.M., Barry K.A., Nelson K.E., Torralba M., Henrissat B., Coutinho P.M., et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2011;5:639–649. doi: 10.1038/ismej.2010.162. - DOI - PMC - PubMed
-
- Hildebrandt M.A., Hoffmann C., Sherrill-Mix S.A., Keilbaugh S.A., Hamady M., Chen Y.Y., Knight R., Ahima R.S., Bushman F., Wu G.D. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724.e2. doi: 10.1053/j.gastro.2009.08.042. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

