Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance
- PMID: 25955967
- PMCID: PMC4450441
- DOI: 10.7554/eLife.07205
Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance
Abstract
Inheritance of gene expression states is fundamental for cells to 'remember' past events, such as environmental or developmental cues. The conserved Polycomb Repressive Complex 2 (PRC2) maintains epigenetic repression of many genes in animals and plants and modifies chromatin at its targets. Histones modified by PRC2 can be inherited through cell division. However, it remains unclear whether this inheritance can direct long-term memory of individual gene expression states (cis memory) or instead if local chromatin states are dictated by the concentrations of diffusible factors (trans memory). By monitoring the expression of two copies of the Arabidopsis Polycomb target gene FLOWERING LOCUS C (FLC) in the same plants, we show that one copy can be repressed while the other is active. Furthermore, this 'mixed' expression state is inherited through many cell divisions as plants develop. These data demonstrate that epigenetic memory of FLC expression is stored not in trans but in cis.
Keywords: FLOWERING LOCUS C; Polycomb; arabidopsis; chromosomes; cis memory; epigenetics; genes; plant biology; vernalization.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

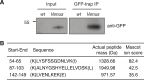

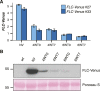


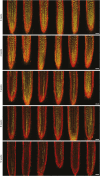

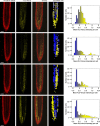
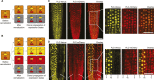
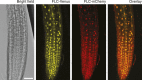

Comment in
-
What holds epigenetic memory?Nat Rev Mol Cell Biol. 2017 Feb 21;18(3):140. doi: 10.1038/nrm.2017.15. Nat Rev Mol Cell Biol. 2017. PMID: 28220047 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

