Novel DNA packaging recognition in the unusual bacteriophage N15
- PMID: 25956737
- PMCID: PMC4461450
- DOI: 10.1016/j.virol.2015.03.027
Novel DNA packaging recognition in the unusual bacteriophage N15
Abstract
Phage lambda's cosB packaging recognition site is tripartite, consisting of 3 TerS binding sites, called R sequences. TerS binding to the critical R3 site positions the TerL endonuclease for nicking cosN to generate cohesive ends. The N15 cos (cos(N15)) is closely related to cos(λ), but whereas the cosB(N15) subsite has R3, it lacks the R2 and R1 sites and the IHF binding site of cosB(λ). A bioinformatic study of N15-like phages indicates that cosB(N15) also has an accessory, remote rR2 site, which is proposed to increase packaging efficiency, like R2 and R1 of lambda. N15 plus five prophages all have the rR2 sequence, which is located in the TerS-encoding 1 gene, approximately 200 bp distal to R3. An additional set of four highly related prophages, exemplified by Monarch, has R3 sequence, but also has R2 and R1 sequences characteristic of cosB-λ. The DNA binding domain of TerS-N15 is a dimer.
Keywords: Bacteriophage; DNA packaging; Terminase; Virus DNA packaging; Virus DNA recognition; Virus assembly; Virus evolution.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
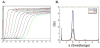


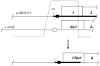
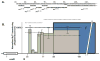

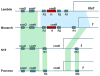
Similar articles
-
The role of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda, in the nicking reaction.J Mol Biol. 1993 Dec 5;234(3):594-609. doi: 10.1006/jmbi.1993.1614. J Mol Biol. 1993. PMID: 8254662
-
Genetic analysis of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage lambda.J Mol Biol. 1992 Nov 5;228(1):58-71. doi: 10.1016/0022-2836(92)90491-2. J Mol Biol. 1992. PMID: 1447794
-
Mutations in Nu1, the gene encoding the small subunit of bacteriophage lambda terminase, suppress the postcleavage DNA packaging defect of cosB mutations.J Bacteriol. 1997 Apr;179(8):2479-85. doi: 10.1128/jb.179.8.2479-2485.1997. J Bacteriol. 1997. PMID: 9098042 Free PMC article.
-
Virus DNA packaging: the strategy used by phage lambda.Mol Microbiol. 1995 Jun;16(6):1075-86. doi: 10.1111/j.1365-2958.1995.tb02333.x. Mol Microbiol. 1995. PMID: 8577244 Review.
-
Viral Small Terminase: A Divergent Structural Framework for a Conserved Biological Function.Viruses. 2022 Oct 8;14(10):2215. doi: 10.3390/v14102215. Viruses. 2022. PMID: 36298770 Free PMC article. Review.
Cited by
-
Enteric Chromosomal Islands: DNA Packaging Specificity and Role of λ-like Helper Phage Terminase.Viruses. 2022 Apr 15;14(4):818. doi: 10.3390/v14040818. Viruses. 2022. PMID: 35458547 Free PMC article.
-
DNA Packaging Specificity of Bacteriophage N15 with an Excursion into the Genetics of a Cohesive End Mismatch.PLoS One. 2015 Dec 3;10(12):e0141934. doi: 10.1371/journal.pone.0141934. eCollection 2015. PLoS One. 2015. PMID: 26633301 Free PMC article.
-
DNA Topology and the Initiation of Virus DNA Packaging.PLoS One. 2016 May 4;11(5):e0154785. doi: 10.1371/journal.pone.0154785. eCollection 2016. PLoS One. 2016. PMID: 27144448 Free PMC article.
References
-
- Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. Journal of molecular biology. 2005;347:895. - PubMed
-
- Arber WELHBMNE, Murray K. Experimental methods for use with lambda. In: Hendrix RWRJWSFW, Weisberg RA, editors. Lambda II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1983. p. 433.
-
- Becker A, Murialdo H, Gold M. Early events in the in vitro packaging of bacteriophage DNA. Virology. 1977;78:291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

