Bacterial Filament Systems: Toward Understanding Their Emergent Behavior and Cellular Functions
- PMID: 25957405
- PMCID: PMC4498058
- DOI: 10.1074/jbc.R115.637876
Bacterial Filament Systems: Toward Understanding Their Emergent Behavior and Cellular Functions
Abstract
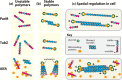
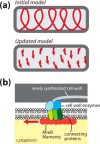
Bacteria use homologs of eukaryotic cytoskeletal filaments to conduct many different tasks, controlling cell shape, division, and DNA segregation. These filaments, combined with factors that regulate their polymerization, create emergent self-organizing machines. Here, we summarize the current understanding of the assembly of these polymers and their spatial regulation by accessory factors, framing them in the context of being dynamical systems. We highlight how comparing the in vivo dynamics of the filaments with those measured in vitro has provided insight into the regulation, emergent behavior, and cellular functions of these polymeric systems.
Keywords: FtsZ; MreB; actin; bacteria; cell biology; cytoskeleton; tubulin.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Erickson H. P. (1995) FtsZ, a prokaryotic homolog of tubulin? Cell 80, 367–370 - PubMed
-
- van den Ent F., Amos L. A., Löwe J. (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44 - PubMed
-
- Margolin W. (1998) A green light for the bacterial cytoskeleton. Trends Microbiol. 6, 233–238 - PubMed
-
- Graumann P. L., Defeu Soufo H. J. (2004) An intracellular actin motor in bacteria? Bioessays 26, 1209–1216 - PubMed
-
- Bi E. F., Lutkenhaus J. (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

