Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain
- PMID: 25960343
- PMCID: PMC4432642
- DOI: 10.1038/ncomms8119
Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain
Abstract
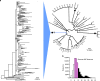
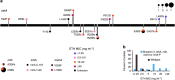
The rise of drug-resistant strains is a major challenge to containing the tuberculosis (TB) pandemic. Yet, little is known about the extent of resistance in early years of chemotherapy and when transmission of resistant strains on a larger scale became a major public health issue. Here we reconstruct the timeline of the acquisition of antimicrobial resistance during a major ongoing outbreak of multidrug-resistant TB in Argentina. We estimate that the progenitor of the outbreak strain acquired resistance to isoniazid, streptomycin and rifampicin by around 1973, indicating continuous circulation of a multidrug-resistant TB strain for four decades. By around 1979 the strain had acquired additional resistance to three more drugs. Our results indicate that Mycobacterium tuberculosis (Mtb) with extensive resistance profiles circulated 15 years before the outbreak was detected, and about one decade before the earliest documented transmission of Mtb strains with such extensive resistance profiles globally.
Figures


References
-
- World Health Organization. ANTIMICROBIAL RESISTANCE Global Report on Surveillance WHO (2014).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

